Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila - PubMed (original) (raw)
Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila
Leah R Sabin et al. Cell. 2009.
Abstract
Intrinsic immune responses autonomously inhibit viral replication and spread. One pathway that restricts viral infection in plants and insects is RNA interference (RNAi), which targets and degrades viral RNA to limit infection. To identify additional genes involved in intrinsic antiviral immunity, we screened Drosophila cells for modulators of viral infection using an RNAi library. We identified Ars2 as a key component of Drosophila antiviral immunity. Loss of Ars2 in cells, or in flies, increases susceptibility to RNA viruses. Consistent with its antiviral properties, we found that Ars2 physically interacts with Dcr-2, modulates its activity in vitro, and is required for siRNA-mediated silencing. Furthermore, we show that Ars2 plays an essential role in miRNA-mediated silencing, interacting with the Microprocessor and stabilizing pri-miRNAs. The identification of Ars2 as a player in these small RNA pathways provides new insight into the biogenesis of small RNAs that may be extended to other systems.
Figures
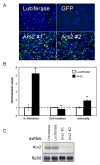
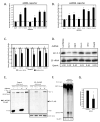
Figure 1. Ars2 controls VSV infection in cell culture
(A) Cells were treated with the indicated dsRNAs; Ars2 #1 and #2 denote independent dsRNA amplicons. Infected cells expressing a VSV-encoded GFP reporter are shown in green, nuclei in blue. (B) Infected Ars2-deficient cells were normalized to infected control cells and percent infection, cell number and average intensity were quantified. * denotes p<1×0-5; error bars indicate +/- SD from a representative experiment. Experiment was repeated 5 times. (C) Northern analysis of cells treated with the indicated dsRNAs.
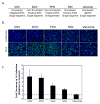
Figure 2. Ars2 controls infection of a panel of RNA viruses in cell culture
(A) Table of the name and the physical properties of each virus. (B) Immunofluorescence of DL1 cells infected with a diverse panel of viruses. Cells were treated with dsRNAs against Ars2 or control. Virus antigen is shown in green, nuclei in blue. (C) Quantification of images as in (B). All values were significant at p<0.0001; error bars indicate +/- SD of three independent experiments.
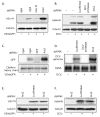
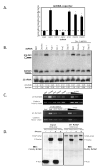
Figure 3. Ars2 controls viral RNA levels during RNA virus infection
(A) Immunoblot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with VSV as indicated. (B) Immunoblot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with DCV as indicated. (C) Northern blot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with VSV as indicated. (D) Northern blot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with DCV as indicated. (E) Entry assay by immunoblot analysis. Cells pre-treated with the indicated dsRNAs were either uninfected or were infected with VSV, extracellular virus was removed, and immunoblot analysis was performed against internalized VSV (VSV M) or control (tubulin). (F) Entry assay by immunoblot analysis. Cells pre-treated with the indicated dsRNAs were either uninfected or were infected with DCV, extracellular virus was removed, and immunoblot analysis was performed against internalized DCV (capsids) or the control (tubulin).
Figure 4. Ars2 is required for antiviral immunity in adult flies
(A) Ars2 is developmentally required in Drosophila. Table of Ars2 mutants and their phenotypes. (B) Adult flies carrying heat shock inducible Gal4 were crossed to flies that can be induced to express dsRNA against Ars2 (hs-Gal4>UAS-Ars2 IR) or controls (hs-Gal4>+). Adult progeny controls (open circles) or Ars2-deficient flies (open triangles) were challenged with vehicle (PBS) and mortality was monitored as a function of time post-injection. In addition, control (closed circles) or Ars2-deficient flies (closed triangles) were challenged with DCV and mortality was monitored as a function of time post-infection. Percent survival is graphed as a function of time (p<0.001 log rank test). A representative of at least three experiments is shown. (C) Immunoblot analysis of flies that are either depleted for Ars2 (mut) or controls (wt) as described in (B), infected with DCV and collected at the indicated time points post infection. (D) Ars2-deficient flies (triangles) or controls (circles) as described in (B) were challenged with VSV and monitored daily for mortality; percent survival is graphed as a function of time (p<0.001 log rank test). A representative of at least three experiments is shown. (E) Immunoblot analysis of flies that are either depleted for Ars2 (mut) or controls (wt) as described in (B), infected with VSV and collected at the indicated time points post infection.
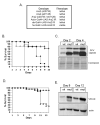
Figure 5. Ars2 is required for siRNA-mediated silencing
(A) Cells were transfected with a control reporter (Firefly), a Renilla reporter, and a vector expressing a Renilla hairpin. The indicated proteins were depleted by RNAi and cells were monitored for siRNA-mediated silencing. * denotes p<1×0-5. (B) Cells were transfected with a control vector (Firefly) and Renilla-mus308. The indicated proteins were depleted by RNAi and the cells were monitored for esiRNA-mediated silencing. * denotes p<1×0-5. (C) Cells were transfected as in (A) however siRNA duplexes against Renilla were transfected in lieu of the long hairpin trigger vector. * denotes p<0.001. (D) Northern blot analysis of esi-2.1 in cells depleted as indicated. esi-2.1 expression was quantified relative to the 2S rRNA, and values were normalized to the Luciferase negative control. (E) Cells expressing NTAP-Ars2 and either FLAG-Ran or FLAG-Dcr-2 were immunoprecipitated, treated with RNase as indicated, and immuoblotted for FLAG. Left panel shows 1/10 input and the right panel shows the co-immunoprecipitate. (F) Cytoplasmic extracts from cells expressing FLAG-Dcr-2 were depleted for Ars2 or Luciferase control (Con), and incubated with a uniformly radiolabeled dsRNA substrate to measure siRNA-generating activity. (G) Quantification of siRNA production normalized to the siRNA-generating activity of the control extract. * denotes p<0.05. Error bars represent +/- SD of three independent experiments.
Figure 6. Ars2 is required for miRNA-mediated silencing
(A) Cells were transfected with a control reporter (Firefly), a vector expressing pri-miR-277, and a Renilla reporter bearing bulged miR-277 target sites. The indicated proteins were depleted by RNAi and the cells were monitored for miRNA-mediated silencing. * denotes p<0.0001; error bars represent +/- SD of 3 independent experiments. (B) Northern blot analysis of pre- and mature bantam miRNA. Expression of pre- and mature bantam was quantified relative to the 2S rRNA, and values were normalized to βgal. (C) RT-PCR analysis of pri-bantam and cellular control transcript (clathrin heavy chain) levels in cells depleted as indicated. Serial five-fold dilutions indicate amplification in the linear range. (D) Cells expressing NTAP-Ars2 and either FLAG-Ran or FLAG-Pasha were immunoprecipitated, treated with RNase as indicated, and immuoblotted for FLAG. Left panel shows the 1/10 input and the right panel shows the co-immunoprecipitate.
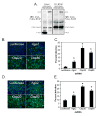
Figure 7. The CBC interacts with Ars2 and restricts RNA virus infection
(A) Cells expressing NTAP-Cbp20 and either control (FLAG-Ran) or Ars2 (pMT-Gal4>UAS-Ars2) were immunoprecipitated and immunoblotted for FLAG and Ars2. Left panel shows the 1/10 input and the right panel shows the co-immunoprecipitate. * denotes nonspecific cross-reacting proteins. (B) Fluorescence of cells pre-treated with dsRNAs against the indicated genes and infected with VSV. Infected cells expressing GFP are shown in green, nuclei in blue. (C) Percent infection was calculated by dividing the number of GFP-positive cells by the total number of cells. * denotes p<1×0-8. (D) Immunofluorescence of cells pre-treated with dsRNAs against the indicated genes and infected with DCV. Infected cells expressing capsids are shown in green, nuclei in blue. (E) Percent infection was calculated by dividing the number of DCV-positive cells by the total number of cells. * denotes p<1×0-8. Error bars represent +/- SD of three independent experiments.
Comment in
- Ars2 and the Cap-Binding Complex Team up for Silencing.
Nielsen AF, Gloggnitzer J, Martinez J. Nielsen AF, et al. Cell. 2009 Jul 23;138(2):224-6. doi: 10.1016/j.cell.2009.07.009. Cell. 2009. PMID: 19632172
Similar articles
- A DNA virus-encoded immune antagonist fully masks the potent antiviral activity of RNAi in Drosophila.
Bronkhorst AW, Vogels R, Overheul GJ, Pennings B, Gausson-Dorey V, Miesen P, van Rij RP. Bronkhorst AW, et al. Proc Natl Acad Sci U S A. 2019 Nov 26;116(48):24296-24302. doi: 10.1073/pnas.1909183116. Epub 2019 Nov 11. Proc Natl Acad Sci U S A. 2019. PMID: 31712431 Free PMC article. - Fly antiviral RNA silencing and miRNA biogenesis claim ARS2.
Voinnet O. Voinnet O. Cell Host Microbe. 2009 Aug 20;6(2):99-101. doi: 10.1016/j.chom.2009.08.002. Cell Host Microbe. 2009. PMID: 19683674 Review. - Small RNAs tackle large viruses: RNA interference-based antiviral defense against DNA viruses in insects.
Bronkhorst AW, Miesen P, van Rij RP. Bronkhorst AW, et al. Fly (Austin). 2013 Oct-Dec;7(4):216-23. doi: 10.4161/fly.25708. Epub 2013 Aug 23. Fly (Austin). 2013. PMID: 23974177 Free PMC article. - Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells.
Qi N, Zhang L, Qiu Y, Wang Z, Si J, Liu Y, Xiang X, Xie J, Qin CF, Zhou X, Hu Y. Qi N, et al. J Virol. 2012 May;86(10):5763-73. doi: 10.1128/JVI.07229-11. Epub 2012 Mar 21. J Virol. 2012. PMID: 22438534 Free PMC article. - The long and short of antiviral defense: small RNA-based immunity in insects.
Bronkhorst AW, van Rij RP. Bronkhorst AW, et al. Curr Opin Virol. 2014 Aug;7:19-28. doi: 10.1016/j.coviro.2014.03.010. Epub 2014 Apr 15. Curr Opin Virol. 2014. PMID: 24732439 Review.
Cited by
- PRP4KA phosphorylates SERRATE for degradation via 20_S_ proteasome to fine-tune miRNA production in Arabidopsis.
Wang L, Yan X, Li Y, Wang Z, Chhajed S, Shang B, Wang Z, Choi SW, Zhao H, Chen S, Zhang X. Wang L, et al. Sci Adv. 2022 Mar 25;8(12):eabm8435. doi: 10.1126/sciadv.abm8435. Epub 2022 Mar 25. Sci Adv. 2022. PMID: 35333566 Free PMC article. - Menin is required for optimal processing of the microRNA let-7a.
Gurung B, Muhammad AB, Hua X. Gurung B, et al. J Biol Chem. 2014 Apr 4;289(14):9902-8. doi: 10.1074/jbc.M113.520692. Epub 2014 Feb 21. J Biol Chem. 2014. PMID: 24563463 Free PMC article. - Emerging Roles of the Nuclear Cap-Binding Complex in Abiotic Stress Responses.
Daszkowska-Golec A. Daszkowska-Golec A. Plant Physiol. 2018 Jan;176(1):242-253. doi: 10.1104/pp.17.01017. Epub 2017 Nov 15. Plant Physiol. 2018. PMID: 29142023 Free PMC article. Review. - Transcriptomic comparison of Drosophila snRNP biogenesis mutants reveals mutant-specific changes in pre-mRNA processing: implications for spinal muscular atrophy.
Garcia EL, Wen Y, Praveen K, Matera AG. Garcia EL, et al. RNA. 2016 Aug;22(8):1215-27. doi: 10.1261/rna.057208.116. Epub 2016 Jun 6. RNA. 2016. PMID: 27268418 Free PMC article. - Innate antiviral immunity in Drosophila.
Sabin LR, Hanna SL, Cherry S. Sabin LR, et al. Curr Opin Immunol. 2010 Feb;22(1):4-9. doi: 10.1016/j.coi.2010.01.007. Curr Opin Immunol. 2010. PMID: 20137906 Free PMC article. Review.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. - PubMed
- Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367:212–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007451/AI/NIAID NIH HHS/United States
- T32 GM-07229/GM/NIGMS NIH HHS/United States
- T32 GM007229/GM/NIGMS NIH HHS/United States
- R01AI07451/AI/NIAID NIH HHS/United States
- R01 AI074951-01/AI/NIAID NIH HHS/United States
- R01 AI074951/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous