LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model - PubMed (original) (raw)
. 2009 Nov 1;18(21):4022-34.
doi: 10.1093/hmg/ddp346. Epub 2009 Jul 29.
Affiliations
- PMID: 19640926
- PMCID: PMC2758136
- DOI: 10.1093/hmg/ddp346
LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model
Javier Alegre-Abarrategui et al. Hum Mol Genet. 2009.
Abstract
Leucine rich repeat kinase 2 (LRRK2) mutations are the most common genetic cause of Parkinson's disease (PD) although LRRK2 function remains unclear. We report a new role for LRRK2 in regulating autophagy and describe the recruitment of LRRK2 to the endosomal-autophagic pathway and specific membrane subdomains. Using a novel human genomic reporter cellular model, we found LRRK2 to locate to membrane microdomains such as the neck of caveolae, microvilli/filopodia and intraluminal vesicles of multivesicular bodies (MVBs). In human brain and in cultured human cells LRRK2 was present in cytoplasmic puncta corresponding to MVBs and autophagic vacuoles (AVs). Expression of the common R1441C mutation from a genomic DNA construct caused impaired autophagic balance evident by the accumulation of MVBs and large AVs containing incompletely degraded material and increased levels of p62. Furthermore, the R1441C mutation induced the formation of skein-like abnormal MVBs. Conversely, LRRK2 siRNA knockdown increased autophagic activity and prevented cell death caused by inhibition of autophagy in starvation conditions. The work necessitated developing a new, more efficient recombineering strategy, which we termed Sequential insertion of Target with ovErlapping Primers (STEP) to seamlessly fuse the green fluorescent protein-derivative YPet to the human LRRK2 protein in the LRRK2 genomic locus carried by a bacterial artificial chromosome. Taken together our data demonstrate the functional involvement of LRRK2 in the endosomal-autophagic pathway and the recruitment to specific membrane microdomains in a physiological human gene expression model suggesting a novel function for this important PD-related protein.
Figures
Figure 1.
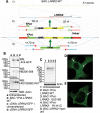
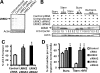
Construction and expression of fluorescently N-terminal tagged genomic fusion constructs of LRRK2 using STEP recombination. (A1) BAC-_LRRK2_-WT containing the LRRK2 human genomic locus was built by combining BAC RP11-115F18 and BAC RP11-568G5 inserts. (A2) In the first stage of STEP recombination a linear cassette containing the selection/counter-selection gene RpsL-neo was inserted in LRRK2 exon 1 at the starting ATG by homologous recombination. The RpsL-neo cassette was created by a single PCR reaction incorporating four 80-mer overlapping primers. The cassette contained at the 3′ and 5′ ends 55 bp homology regions which flanked the first ≈55 bp and last ≈55 bp, respectively, of Ypet and a peptide linker. (A3) The RpsL-neo gene was replaced by a marker-less linear cassette containing the full sequence of Ypet and additional homology regions created again by a single PCR reaction. Homologous recombination occurred efficiently through the extended ≈170 bp homology regions. (B) Protein lysates from cells transfected to express either cDNA-_LRRK2_-GFP or BAC-YPet-_LRRK2_-WT were used for immunoblotting using either an anti-GFP antibody (which recognizes YPet and GFP) or two anti-LRRK2 antibodies. The amount of tagged LRRK2 protein was equalized by loading 1 µg of total protein for cDNA-_LRRK2_-GFP and 100 µg of total protein for BAC-YPet-_LRRK2_-WT. A control was included containing 100 µg of protein from untransfected cells together with 1 µg of lysate containing overexpressed cDNA derived protein. (C) BAC-YPet-_LRRK2_-WT was engineered to carry either the G2019S or the R1441C mutations using recombineering. The N-terminally tagged BAC-YPet-_LRRK2_-WT, BAC-YPet-_LRRK2_-G2019S or BAC-YPet-_LRRK2_-R1441C as well as the C-terminally tagged BAC_-LRRK2-_Ypet-WT expressed full length LRRK2 protein as detected using anti-GFP antibodies. (D and E) Cellular expression of BAC-YPet-_LRRK2_-WT revealed a cytoplasmic pattern with the appearance of fine cytoplasmic puncta in Vero (D) and HEK293 (E) cells as detected by IF using anti-GFP antibodies.
Figure 2.
Appearance of LRRK2 puncta in human brain by IF. Staining of LRRK2 puncta in: (A) A Purkinje cell in a control case; (B) vascular endothelial cells in the striatum of a MSA patient; (C–E) fascia dentata of a patient diagnosed with both AD and PD (C), a 2-month-old infant (D) and an aged control (E). Anti-LRRK2 antibodies NB300-268 (A, B, C and E) or NB300-267 (D) were used.
Figure 3.
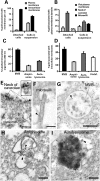
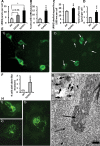
LRRK2 localizes to the neck of caveolae, microvilli and the endosomal–autophagic pathway. (A) Quantification of the distribution of membrane-associated LRRK2 between the plasma membrane and intracellular membranous structures in attached or trypsinized cells. (B) Quantification of the distribution of LRRK2 within distinct plasma membrane domains in attached or trypsinized cells. (C) Quantification of the proportion of different organelles ≥200 nm (the resolution limit of light microscopy) labelled with LRRK2. (D) Quantification of the amount of LRRK2 within distinct intracellular membranous organelles. (E–I) IEM micrographs show LRRK2 gold labelling on the neck of caveolae (E and amplification of region identified by arrowhead in inset), microvilli (F), intraluminar vesicles of MVBs (G), amphisomes (H) and autolysosomes (I). Representative gold particles labelling LRRK2 are identified using arrowheads. HEK293 cells were transfected with BAC-YPet-_LRRK2_-WT and IEM was obtained with anti-GFP antibodies. Quantification of gold particles was obtained by counting eight random labelled cells. Bars represent mean + SEM. Scale bars represent 100 nm.
Figure 4.
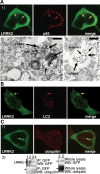
A proportion of LRRK2 puncta colocalize with the proteins p62 and LC3, but not with ubiquitin. (A) LRRK2 puncta colocalize with p62 endogenous puncta. BAC-YPet-_LRRK2_-WT was transfected in HEK-293 cells and double-labelling performed by IF (A1) or IEM (A2 and 3) using anti-GFP (thick arrows in A2 and 3) and either anti-p62 [A1 and 2 (thin arrows in A2)] or anti-annexin1 (A3) antibodies (thin arrows). Scale bar in image A2 represents 200 nm although in A3 represents 500 nm. (B) LRRK2 puncta colocalize with LC3 protein. BAC-YPet-_LRRK2_-WT was transfected in HEK-293 cells stably expressing HA-mCherry-LC3 and analysed by IF using anti-HA and anti-GFP antibodies. (C) BAC-YPet-_LRRK2_-WT, BAC-YPet-_LRRK2_-G2019S or BAC-YPet-_LRRK2_-R1441C were expressed in transfected cells and double labelling performed using YPet-LRRK2 fluorescence and anti-ubiquitin antibodies (BAC-YPet-_LRRK2_-WT in C1). Alternatively protein lysates were prepared and subjected to immunoblotting either directly or after immunoprecipitation using anti-GFP antibodies (C2). Legend of samples: 1 = Untransfected, 2 = BAC-YPet_-LRRK2_-WT, 3 = BAC-YPet-_LRRK2_-G2019S and 4 = BAC-YPet-_LRRK2_-R1441C. A1, B and C1 are confocal images.
Figure 5.
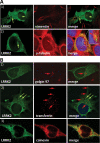
LRRK2 puncta do not colocalize with aggresome markers. LRRK2 shows a discrete colocalization with calnexin (ER) but not with the Golgi apparatus (golgin97) or recycling endosomes (transferrin). (A) BAC-YPet-_LRRK2_-WT was expressed in transfected cells and IF performed using anti-GFP and either anti-vimentin (A1) or anti-γ-tubulin antibodies (A2). LRRK2 puncta frequently located close to the γ-tubulin positive centrosome (A2 inset). (B) Cells transfected with BAC-YPet-_LRRK2_-WT were subjected to IF with anti-GFP and either anti-golgin97 (TGN) or calnexin (ER) antibodies (B1 and 3). Alternatively cells were loaded with alexa-594-transferrin (recycling endosomes) and IF performed with anti-GFP detected by alexa-488 conjugated secondary antibodies (B2).
Figure 6.
LRRK2 knockdown increases autophagic activity and prevents Bafilomycin-induced cell death under starvation. (A) Demonstration of siRNA mediated LRRK2 knockdown. siRNA mediated knockdown of wild-type Ypet-LRRK2 in clonal cell lines carrying BAC-YPet-_LRRK2_-WT-FRT shown by western blot using anti-GFP antibodies. Forty micrograms of protein were loaded per lane. (B) Upon LRRK2 knockdown, LC3-I and LC3-II endogenous expression was assessed by western blot under nutrient-rich or 4.5 h starvation in HBSS with or without 200 nM of the autophagic inhibitor bafilomycin A1 (BFA). Ten micrograms of protein were loaded per lane. Mean LC3-II densitometric relative values versus actin of three experiments are shown. (C) Quantification of LC3-II turnover under starvation upon LRRK2 siRNA knockdown after normalization against actin using a densitometric analysis. The amount of LC3-II under starvation was subtracted from the amount of LC3-II under starvation in the presence of the inhibitor of late stages of autophagy BFA, and the difference expressed as a percentage relative to the amount of LC3-II under starvation in the presence of BFA. (D) Quantification of the relative amount of cells per well surviving 4.5 h of starvation treatment in HBSS with or without BFA upon LRRK2 knockdown with respect to nutrient rich conditions. Values are expressed as percentages against the values obtained from cells cultured in normal media for each condition. Bars represent mean + SEM. Experiments were performed in triplicate. Statistical comparisons are made against the control using Student's t test; *P < 0.05, **P < 0.01.
Figure 7.
LRRK2 pathogenic mutations increase the number and size of LRRK2 puncta and induce the formation of skein-like structures when expressed from the genomic DNA constructs. (A) Expression of the R1441C LRRK2 pathogenic mutation from BAC-YPet-_LRRK2_-R1441C showed a ≈3-fold increase respect to WT in the number of LRRK2 puncta per cell (Vero) assessed by IF using anti-GFP antibodies (P < 0.05, Fisher's exact test). Puncta in 20 random labelled cells per genotype were counted using a 63× objective. The G2019G LRRK2 mutation showed a trend of a ≈2-fold increase which did not reach statistical significance. (**B**) The R1441C mutation also showed a ≈3-fold increase in the percentage of cells (HEK293) harbouring observable LRRK2 puncta assessed by IF (_P_ = 0.0001, Fisher's exact test), illustrated in (**E**) [E1 (WT) and E2 (R1441C)]. (**C**) Quantification of the number of labelled organelles >200 nm (the resolution limit of light microscopy) assessed by IEM. (D) The R1441C induced a ≈2-fold increase in the total area per cell occupied by AVs assessed by IEM. The total cell area remained constant between genotypes. (F) The R1441C LRRK2 pathogenic mutation led to the formation of skein-like three dimensional structures in a statistically significant number of cells (P = 0.0038, Fisher's exact test) (HEK293 cells) which are illustrated in z-stack IF pictures H1 to 3 (anti-GFP antibodies). (H) The R1441C LRRK2 skein-like structures were frequently interspersed with LRRK2 puncta and pointed towards a paranuclear location. (G) The skein-like structures appeared under the electron microscope as membrane (arrowheads) contained structures heavily positive for LRRK2 (anti-GFP antibodies) and composed of a densely packed core (thick arrow and amplification in inset) and a fine granular matrix (thin arrow). Scale bar represents 2 µm. Six randomly-chosen fields of view per genotype where analysed using a 20× objective in IF quantifications except where otherwise indicated. Eight randomly-chosen labelled cells per genotype were analysed in IEM quantifications. Counting of all structures was done blind to the genotype. Bars represent mean + SEM. Statistical significances were obtained using a Student's t test except where indicated; *P < 0.05. **P < 0.01.
Similar articles
- Parkinson disease, LRRK2 and the endocytic-autophagic pathway.
Alegre-Abarrategui J, Wade-Martins R. Alegre-Abarrategui J, et al. Autophagy. 2009 Nov;5(8):1208-10. doi: 10.4161/auto.5.8.9894. Epub 2009 Nov 24. Autophagy. 2009. PMID: 19770575 - 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization.
Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, Alessi DR. Nichols RJ, et al. Biochem J. 2010 Sep 15;430(3):393-404. doi: 10.1042/BJ20100483. Biochem J. 2010. PMID: 20642453 Free PMC article. - Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells.
Plowey ED, Cherra SJ 3rd, Liu YJ, Chu CT. Plowey ED, et al. J Neurochem. 2008 May;105(3):1048-56. doi: 10.1111/j.1471-4159.2008.05217.x. Epub 2008 Jan 7. J Neurochem. 2008. PMID: 18182054 Free PMC article. - Genetic analysis of Parkinson's disease-linked leucine-rich repeat kinase 2.
Tong Y, Shen J. Tong Y, et al. Biochem Soc Trans. 2012 Oct;40(5):1042-6. doi: 10.1042/BST20120112. Biochem Soc Trans. 2012. PMID: 22988862 Free PMC article. Review. - LRRK2 and autophagy: a common pathway for disease.
Manzoni C. Manzoni C. Biochem Soc Trans. 2012 Oct;40(5):1147-51. doi: 10.1042/BST20120126. Biochem Soc Trans. 2012. PMID: 22988880 Review.
Cited by
- LRRK2 Pathways Leading to Neurodegeneration.
Cookson MR. Cookson MR. Curr Neurol Neurosci Rep. 2015 Jul;15(7):42. doi: 10.1007/s11910-015-0564-y. Curr Neurol Neurosci Rep. 2015. PMID: 26008812 Free PMC article. Review. - GTPase activity and neuronal toxicity of Parkinson's disease-associated LRRK2 is regulated by ArfGAP1.
Stafa K, Trancikova A, Webber PJ, Glauser L, West AB, Moore DJ. Stafa K, et al. PLoS Genet. 2012;8(2):e1002526. doi: 10.1371/journal.pgen.1002526. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22363216 Free PMC article. - Selective expression of Parkinson's disease-related Leucine-rich repeat kinase 2 G2019S missense mutation in midbrain dopaminergic neurons impairs dopamine release and dopaminergic gene expression.
Liu G, Sgobio C, Gu X, Sun L, Lin X, Yu J, Parisiadou L, Xie C, Sastry N, Ding J, Lohr KM, Miller GW, Mateo Y, Lovinger DM, Cai H. Liu G, et al. Hum Mol Genet. 2015 Sep 15;24(18):5299-312. doi: 10.1093/hmg/ddv249. Epub 2015 Jun 29. Hum Mol Genet. 2015. PMID: 26123485 Free PMC article. - Leucine-rich repeat kinase 2 for beginners: six key questions.
Kett LR, Dauer WT. Kett LR, et al. Cold Spring Harb Perspect Med. 2012 Mar;2(3):a009407. doi: 10.1101/cshperspect.a009407. Cold Spring Harb Perspect Med. 2012. PMID: 22393539 Free PMC article. Review. - Comprehensive Genomic Analysis Reveals the Prognostic Role of LRRK2 Copy-Number Variations in Human Malignancies.
Lopez G, Lazzeri G, Rappa A, Isimbaldi G, Cribiù FM, Guerini-Rocco E, Ferrero S, Vaira V, Di Fonzo A. Lopez G, et al. Genes (Basel). 2020 Jul 24;11(8):846. doi: 10.3390/genes11080846. Genes (Basel). 2020. PMID: 32722212 Free PMC article.
References
- Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. - PubMed
- Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. - PubMed
- Mata I.F., Kachergus J.M., Taylor J.P., Lincoln S., Aasly J., Lynch T., Hulihan M.M., Cobb S.A., Wu R.M., Lu C.S., et al. Lrrk2 pathogenic substitutions in Parkinson's disease. Neurogenetics. 2005;6:171–177. - PubMed
- Mata I.F., Wedemeyer W.J., Farrer M.J., Taylor J.P., Gallo K.A. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends. Neurosci. 2006;29:286–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases