O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress - PubMed (original) (raw)
Review
O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress
Chutikarn Butkinaree et al. Biochim Biophys Acta. 2010 Feb.
Abstract
Background: Since its discovery in the early 1980s, O-linked-beta-N-acetylglucosamine (O-GlcNAc), a single sugar modification on the hydroxyl group of serine or threonine residues, has changed our views of protein glycosylation. While other forms of protein glycosylation modify proteins on the cell surface or within luminal compartments of the secretory machinery, O-GlcNAc modifies myriad nucleocytoplasmic proteins. GlcNAcylated proteins are involved in transcription, ubiquitination, cell cycle, and stress responses. GlcNAcylation is similar to protein phosphorylation in terms of stoichiometry, localization and cycling. To date, only two enzymes are known to regulate GlcNAcylation in mammals: O-GlcNAc transferase (OGT), which catalyzes the addition of O-GlcNAc, and beta-N-acetylglucosaminidase (O-GlcNAcase), a neutral hexosaminidase responsible for O-GlcNAc removal. OGT and O-GlcNAcase are regulated by RNA splicing, by nutrients, and by post-translational modifications. Their specificities are controlled by many transiently associated targeting subunits. As methods for detecting O-GlcNAc have improved our understanding of O-GlcNAc's functions has grown rapidly.
Scope of review: In this review, the functions of GlcNAcylation in regulating cellular processes, its extensive crosstalk with protein phosphorylation, and regulation of OGT and O-GlcNAcase will be explored.
Major conclusions: GlcNAcylation rivals phosphorylation in terms of its abundance, protein distribution and its cycling on and off of proteins. GlcNAcylation has extensive crosstalk with phosphorylation to regulate signaling, transcription and the cytoskeleton in response to nutrients and stress.
General significance: Abnormal crosstalk between GlcNAcylation and phosphorylation underlies dysregulation in diabetes, including glucose toxicity, and defective GlcNAcylation is involved in neurodegenerative disease and cancer and most recently in AIDS.
Copyright 2009 Elsevier B.V. All rights reserved.
Figures
Figure 1
Hexosamine biosynthetic pathway (HBP). The synthesis of UDP-GlcNAc from glucose and enzymes involved in the process are shown. Commonly used inhibitors of HBP and GlcNAcylation are shown in red.
Figure 2
Schematic structures of OGT and O-GlcNAcase. A. Three different forms of human OGT are produced by alternative splicing from a single gene which resides near the gene of Parkinson's dystonia in chromosome X. Known post-translational modifications are indicated by amino acid positions (if available). B. Two isoforms of O-GlcNAcase are known. The gene is located near a locus of late on-set Alzheimer's disease. Known post-translational modifications are indicated by amino acid positions and a caspase-3 cleavage site is shown. NLS: nuclear localization signal, MLS: mitochondrial localization signal.
Figure 3
Mechanism of OGT regulation. OGT is regulated by multiple factors including transcriptional regulation, mRNA splicing, donor substrate availability, post-translational modification and multimerization. Multimerized ncOGTs form dynamic holoenzymes with many different protein partners and regulate differential targeting of proteins as well as their GlcNAcylation.
Figure 4
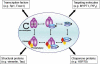
Complex regulation of OGT and O-GlcNAcase. OGT and O-GlcNAcase can form very dynamic complexes with each other and/or kinases and phosphatases under different cellular signalings. Transient complex formation between O-GlcNAc regulatory enzymes and various binding partners enables a highly sensitive regulation of GlcNAcylation in response to specific cellular conditions. OGT: purple, O-GlcNAcase: blue, Kinase: red, Phosphatase: yellow.
Figure 5
Dynamic interplay between of GlcNAcylation and phosphorylation. A. O-GlcNAc and O-phosphate can compete for the same site. This competition can change the activity or stability of the proteins (e.g. c-Myc; Thr 58). B. In some cases, O-GlcNAc and O-phostate modification occurs within ∼10 amino acids range, regulating the function of the protein substrates (e.g. CTD repeat; Ser 2 and 5 for O-phosphate, Thr 4 for O-GlcNAc). C. GlcNAcylation and phosphorylation can occur on the same protein at proximal sites. The balance between GlcNAcylation and phosphorylation can change the cellular function of the protein (e.g. Akt; Thr 308 and Ser 473 for O-phosphate, Ser 473 for O-GlcNAc).
Similar articles
- Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity.
Copeland RJ, Bullen JW, Hart GW. Copeland RJ, et al. Am J Physiol Endocrinol Metab. 2008 Jul;295(1):E17-28. doi: 10.1152/ajpendo.90281.2008. Epub 2008 Apr 29. Am J Physiol Endocrinol Metab. 2008. PMID: 18445751 Free PMC article. Review. - Nutrient regulation of signaling and transcription.
Hart GW. Hart GW. J Biol Chem. 2019 Feb 15;294(7):2211-2231. doi: 10.1074/jbc.AW119.003226. Epub 2019 Jan 9. J Biol Chem. 2019. PMID: 30626734 Free PMC article. Review. - Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation.
Hu P, Shimoji S, Hart GW. Hu P, et al. FEBS Lett. 2010 Jun 18;584(12):2526-38. doi: 10.1016/j.febslet.2010.04.044. Epub 2010 Apr 22. FEBS Lett. 2010. PMID: 20417205 Review. - Feedback Regulation of _O_-GlcNAc Transferase through Translation Control to Maintain Intracellular _O_-GlcNAc Homeostasis.
Lin CH, Liao CC, Chen MY, Chou TY. Lin CH, et al. Int J Mol Sci. 2021 Mar 27;22(7):3463. doi: 10.3390/ijms22073463. Int J Mol Sci. 2021. PMID: 33801653 Free PMC article. - The Beginner's Guide to _O_-GlcNAc: From Nutrient Sensitive Pathway Regulation to Its Impact on the Immune System.
Mannino MP, Hart GW. Mannino MP, et al. Front Immunol. 2022 Jan 31;13:828648. doi: 10.3389/fimmu.2022.828648. eCollection 2022. Front Immunol. 2022. PMID: 35173739 Free PMC article. Review.
Cited by
- O-GlcNAcylation of a circadian clock protein: dPER taking its sweet time.
Diernfellner AC, Brunner M. Diernfellner AC, et al. Genes Dev. 2012 Mar 1;26(5):415-6. doi: 10.1101/gad.188524.112. Genes Dev. 2012. PMID: 22391445 Free PMC article. - Differential effects of an O-GlcNAcase inhibitor on tau phosphorylation.
Yu Y, Zhang L, Li X, Run X, Liang Z, Li Y, Liu Y, Lee MH, Grundke-Iqbal I, Iqbal K, Vocadlo DJ, Liu F, Gong CX. Yu Y, et al. PLoS One. 2012;7(4):e35277. doi: 10.1371/journal.pone.0035277. Epub 2012 Apr 19. PLoS One. 2012. PMID: 22536363 Free PMC article. - Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-κB.
Phoomak C, Vaeteewoottacharn K, Sawanyawisuth K, Seubwai W, Wongkham C, Silsirivanit A, Wongkham S. Phoomak C, et al. Sci Rep. 2016 Jun 13;6:27853. doi: 10.1038/srep27853. Sci Rep. 2016. PMID: 27290989 Free PMC article. - Protein O-GlcNAcylation Promotes Trophoblast Differentiation at Implantation.
Ruane PT, Tan CMJ, Adlam DJ, Kimber SJ, Brison DR, Aplin JD, Westwood M. Ruane PT, et al. Cells. 2020 Oct 6;9(10):2246. doi: 10.3390/cells9102246. Cells. 2020. PMID: 33036308 Free PMC article. - Mass spectrometry based glycoproteomics--from a proteomics perspective.
Pan S, Chen R, Aebersold R, Brentnall TA. Pan S, et al. Mol Cell Proteomics. 2011 Jan;10(1):R110.003251. doi: 10.1074/mcp.R110.003251. Epub 2010 Aug 24. Mol Cell Proteomics. 2011. PMID: 20736408 Free PMC article. Review.
References
- Torres CR, Hart GW. J Biol Chem. 1984;259:3308–17. - PubMed
- Holt GD, Haltiwanger RS, Torres CR, Hart GW. J Biol Chem. 1987;262:14847–50. - PubMed
- Holt GD, Hart GW. J Biol Chem. 1986;261:8049–57. - PubMed
- Bouche C, Serdy S, Kahn CR, Goldfine AB. Endocr Rev. 2004;25:807–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- N01HV28180/HV/NHLBI NIH HHS/United States
- R01 CA042486-26/CA/NCI NIH HHS/United States
- CA42486/CA/NCI NIH HHS/United States
- R01 CA042486/CA/NCI NIH HHS/United States
- R33 DK071280-04/DK/NIDDK NIH HHS/United States
- R33 DK071280/DK/NIDDK NIH HHS/United States
- R01 DK061671/DK/NIDDK NIH HHS/United States
- R37 HD013563-30/HD/NICHD NIH HHS/United States
- DK61671/DK/NIDDK NIH HHS/United States
- N01-HV-28180/HV/NHLBI NIH HHS/United States
- R37 HD013563/HD/NICHD NIH HHS/United States
- R01 DK061671-07/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous