A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2 - PubMed (original) (raw)
. 2009 Sep;41(9):996-1000.
doi: 10.1038/ng.424. Epub 2009 Aug 2.
Susan J Ramus, Jonathan Tyrer, Kelly L Bolton, Aleksandra Gentry-Maharaj, Eva Wozniak, Hoda Anton-Culver, Jenny Chang-Claude, Daniel W Cramer, Richard DiCioccio, Thilo Dörk, Ellen L Goode, Marc T Goodman, Joellen M Schildkraut, Thomas Sellers, Laura Baglietto, Matthias W Beckmann, Jonathan Beesley, Jan Blaakaer, Michael E Carney, Stephen Chanock, Zhihua Chen, Julie M Cunningham, Ed Dicks, Jennifer A Doherty, Matthias Dürst, Arif B Ekici, David Fenstermacher, Brooke L Fridley, Graham Giles, Martin E Gore, Immaculata De Vivo, Peter Hillemanns, Claus Hogdall, Estrid Hogdall, Edwin S Iversen, Ian J Jacobs, Anna Jakubowska, Dong Li, Jolanta Lissowska, Jan Lubiński, Galina Lurie, Valerie McGuire, John McLaughlin, Krzysztof Medrek, Patricia G Moorman, Kirsten Moysich, Steven Narod, Catherine Phelan, Carole Pye, Harvey Risch, Ingo B Runnebaum, Gianluca Severi, Melissa Southey, Daniel O Stram, Falk C Thiel, Kathryn L Terry, Ya-Yu Tsai, Shelley S Tworoger, David J Van Den Berg, Robert A Vierkant, Shan Wang-Gohrke, Penelope M Webb, Lynne R Wilkens, Anna H Wu, Hannah Yang, Wendy Brewster, Argyrios Ziogas; Australian Cancer (Ovarian) Study; Australian Ovarian Cancer Study Group; Ovarian Cancer Association Consortium; Richard Houlston, Ian Tomlinson, Alice S Whittemore, Mary Anne Rossing, Bruce A J Ponder, Celeste Leigh Pearce, Roberta B Ness, Usha Menon, Susanne Krüger Kjaer, Jacek Gronwald, Montserrat Garcia-Closas, Peter A Fasching, Douglas F Easton, Georgia Chenevix-Trench, Andrew Berchuck, Paul D P Pharoah, Simon A Gayther
Affiliations
- PMID: 19648919
- PMCID: PMC2844110
- DOI: 10.1038/ng.424
A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2
Honglin Song et al. Nat Genet. 2009 Sep.
Abstract
Epithelial ovarian cancer has a major heritable component, but the known susceptibility genes explain less than half the excess familial risk. We performed a genome-wide association study (GWAS) to identify common ovarian cancer susceptibility alleles. We evaluated 507,094 SNPs genotyped in 1,817 cases and 2,353 controls from the UK and approximately 2 million imputed SNPs. We genotyped the 22,790 top ranked SNPs in 4,274 cases and 4,809 controls of European ancestry from Europe, USA and Australia. We identified 12 SNPs at 9p22 associated with disease risk (P < 10(-8)). The most significant SNP (rs3814113; P = 2.5 x 10(-17)) was genotyped in a further 2,670 ovarian cancer cases and 4,668 controls, confirming its association (combined data odds ratio (OR) = 0.82, 95% confidence interval (CI) 0.79-0.86, P(trend) = 5.1 x 10(-19)). The association differs by histological subtype, being strongest for serous ovarian cancers (OR 0.77, 95% CI 0.73-0.81, P(trend) = 4.1 x 10(-21)).
Figures
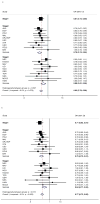
Figure 1
Genotype-specific risks of SNP rs3814113 for ovarian cancer by stage and by study - (a) all epithelial ovarian cancer cases, (b) serous ovarian cancer cases only. Results are based on analyses restricted to subjects of European ancestry.
Figure 2
Map of associated regions on 9p22.2 (nucl. 16655021-17155021). The top panel shows the LD blocks around rs3814113 (±250 kb) for SNPs with MAF≥ 0.05 based on Hapmap CEU data. Squares on the LD block indicate the correlation (r2) between SNPs on a greyscale (darker squares = higher correlations). Approximate location of BNC2 (nucl. 16860786-16399502), CNTLN (nucl. 17125038-17493915) and LOC648570 (nucl. 16777433-16775576) were inferred from the NCBI reference assembly (solid line). The lower panel shows the highlighted LD block in greater detail. The star indicates the location of the most strongly associated SNP rs3814113. The black circles show the position of the other 11 SNPs that reached genome wide significance. The numbers refer to the SNP identifiers in Supplementary Table 2.
Similar articles
- Replication study for the association of seven genome- GWAS-identified Loci with susceptibility to ovarian cancer in the Polish population.
Mostowska A, Sajdak S, Pawlik P, Markowska J, Pawałowska M, Lianeri M, Jagodzinski PP. Mostowska A, et al. Pathol Oncol Res. 2015 Apr;21(2):307-13. doi: 10.1007/s12253-014-9822-6. Epub 2014 Aug 31. Pathol Oncol Res. 2015. PMID: 25173882 Free PMC article. - Genetic variation at 9p22.2 and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers.
Ramus SJ, Kartsonaki C, Gayther SA, Pharoah PD, Sinilnikova OM, Beesley J, Chen X, McGuffog L, Healey S, Couch FJ, Wang X, Fredericksen Z, Peterlongo P, Manoukian S, Peissel B, Zaffaroni D, Roversi G, Barile M, Viel A, Allavena A, Ottini L, Papi L, Gismondi V, Capra F, Radice P, Greene MH, Mai PL, Andrulis IL, Glendon G, Ozcelik H; OCGN; Thomassen M, Gerdes AM, Kruse TA, Cruger D, Jensen UB, Caligo MA, Olsson H, Kristoffersson U, Lindblom A, Arver B, Karlsson P, Stenmark Askmalm M, Borg A, Neuhausen SL, Ding YC, Nathanson KL, Domchek SM, Jakubowska A, Lubiński J, Huzarski T, Byrski T, Gronwald J, Górski B, Cybulski C, Dębniak T, Osorio A, Durán M, Tejada MI, Benítez J, Hamann U, Rookus MA, Verhoef S, Tilanus-Linthorst MA, Vreeswijk MP, Bodmer D, Ausems MG, van Os TA, Asperen CJ, Blok MJ, Meijers-Heijboer HE; HEBON; EMBRACE; Peock S, Cook M, Oliver C, Frost D, Dunning AM, Evans DG, Eeles R, Pichert G, Cole T, Hodgson S, Brewer C, Morrison PJ, Porteous M, Kennedy MJ, Rogers MT, Side LE, Donaldson A, Gregory H, Godwin A, Stoppa-Lyonnet D, Moncoutier V, Castera L, Mazoyer S, Barjhoux L, Bonadona V, Leroux D, Faivre L, Lidereau R, Nogues C, Bignon YJ, Prieur F, Collonge-Rame MA, Venat-… See abstract for full author list ➔ Ramus SJ, et al. J Natl Cancer Inst. 2011 Jan 19;103(2):105-16. doi: 10.1093/jnci/djq494. Epub 2010 Dec 17. J Natl Cancer Inst. 2011. PMID: 21169536 Free PMC article. - Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus.
Zeng C, Guo X, Long J, Kuchenbaecker KB, Droit A, Michailidou K, Ghoussaini M, Kar S, Freeman A, Hopper JL, Milne RL, Bolla MK, Wang Q, Dennis J, Agata S, Ahmed S, Aittomäki K, Andrulis IL, Anton-Culver H, Antonenkova NN, Arason A, Arndt V, Arun BK, Arver B, Bacot F, Barrowdale D, Baynes C, Beeghly-Fadiel A, Benitez J, Bermisheva M, Blomqvist C, Blot WJ, Bogdanova NV, Bojesen SE, Bonanni B, Borresen-Dale AL, Brand JS, Brauch H, Brennan P, Brenner H, Broeks A, Brüning T, Burwinkel B, Buys SS, Cai Q, Caldes T, Campbell I, Carpenter J, Chang-Claude J, Choi JY, Claes KB, Clarke C, Cox A, Cross SS, Czene K, Daly MB, de la Hoya M, De Leeneer K, Devilee P, Diez O, Domchek SM, Doody M, Dorfling CM, Dörk T, Dos-Santos-Silva I, Dumont M, Dwek M, Dworniczak B, Egan K, Eilber U, Einbeigi Z, Ejlertsen B, Ellis S, Frost D, Lalloo F; EMBRACE; Fasching PA, Figueroa J, Flyger H, Friedlander M, Friedman E, Gambino G, Gao YT, Garber J, García-Closas M, Gehrig A, Damiola F, Lesueur F, Mazoyer S, Stoppa-Lyonnet D; behalf of GEMO Study Collaborators; Giles GG, Godwin AK, Goldgar DE, González-Neira A, Greene MH, Guénel P, Haeberle L, Haiman CA, Hallberg E, Hamann U, Hansen TV, Hart S, Hartikainen JM, Ha… See abstract for full author list ➔ Zeng C, et al. Breast Cancer Res. 2016 Jun 21;18(1):64. doi: 10.1186/s13058-016-0718-0. Breast Cancer Res. 2016. PMID: 27459855 Free PMC article. - Functional Analysis and Fine Mapping of the 9p22.2 Ovarian Cancer Susceptibility Locus.
Buckley MA, Woods NT, Tyrer JP, Mendoza-Fandiño G, Lawrenson K, Hazelett DJ, Najafabadi HS, Gjyshi A, Carvalho RS, Lyra PC Jr, Coetzee SG, Shen HC, Yang AW, Earp MA, Yoder SJ, Risch H, Chenevix-Trench G, Ramus SJ, Phelan CM, Coetzee GA, Noushmehr H, Hughes TR, Sellers TA, Goode EL, Pharoah PD, Gayther SA, Monteiro ANA; Ovarian Cancer Association Consortium. Buckley MA, et al. Cancer Res. 2019 Feb 1;79(3):467-481. doi: 10.1158/0008-5472.CAN-17-3864. Epub 2018 Nov 28. Cancer Res. 2019. PMID: 30487138 Free PMC article. - Association between invasive ovarian cancer susceptibility and 11 best candidate SNPs from breast cancer genome-wide association study.
Song H, Ramus SJ, Kjaer SK, DiCioccio RA, Chenevix-Trench G, Pearce CL, Hogdall E, Whittemore AS, McGuire V, Hogdall C, Blaakaer J, Wu AH, Van Den Berg DJ, Stram DO, Menon U, Gentry-Maharaj A, Jacobs IJ, Webb PM, Beesley J, Chen X; Australian Cancer (Ovarian) Study; Australian Ovarian Cancer Study Group; Rossing MA, Doherty JA, Chang-Claude J, Wang-Gohrke S, Goodman MT, Lurie G, Thompson PJ, Carney ME, Ness RB, Moysich K, Goode EL, Vierkant RA, Cunningham JM, Anderson S, Schildkraut JM, Berchuck A, Iversen ES, Moorman PG, Garcia-Closas M, Chanock S, Lissowska J, Brinton L, Anton-Culver H, Ziogas A, Brewster WR, Ponder BA, Easton DF, Gayther SA, Pharoah PD; Ovarian Cancer Association Consortium (OCAC). Song H, et al. Hum Mol Genet. 2009 Jun 15;18(12):2297-304. doi: 10.1093/hmg/ddp138. Epub 2009 Mar 20. Hum Mol Genet. 2009. PMID: 19304784 Free PMC article.
Cited by
- Integrative multi-omics analyses to identify the genetic and functional mechanisms underlying ovarian cancer risk regions.
Dareng EO, Coetzee SG, Tyrer JP, Peng PC, Rosenow W, Chen S, Davis BD, Dezem FS, Seo JH, Nameki R, Reyes AL, Aben KKH, Anton-Culver H, Antonenkova NN, Aravantinos G, Bandera EV, Beane Freeman LE, Beckmann MW, Beeghly-Fadiel A, Benitez J, Bernardini MQ, Bjorge L, Black A, Bogdanova NV, Bolton KL, Brenton JD, Budzilowska A, Butzow R, Cai H, Campbell I, Cannioto R, Chang-Claude J, Chanock SJ, Chen K, Chenevix-Trench G; AOCS Group; Chiew YE, Cook LS, DeFazio A, Dennis J, Doherty JA, Dörk T, du Bois A, Dürst M, Eccles DM, Ene G, Fasching PA, Flanagan JM, Fortner RT, Fostira F, Gentry-Maharaj A, Giles GG, Goodman MT, Gronwald J, Haiman CA, Håkansson N, Heitz F, Hildebrandt MAT, Høgdall E, Høgdall CK, Huang RY, Jensen A, Jones ME, Kang D, Karlan BY, Karnezis AN, Kelemen LE, Kennedy CJ, Khusnutdinova EK, Kiemeney LA, Kjaer SK, Kupryjanczyk J, Labrie M, Lambrechts D, Larson MC, Le ND, Lester J, Li L, Lubiński J, Lush M, Marks JR, Matsuo K, May T, McLaughlin JR, McNeish IA, Menon U, Missmer S, Modugno F, Moffitt M, Monteiro AN, Moysich KB, Narod SA, Nguyen-Dumont T, Odunsi K, Olsson H, Onland-Moret NC, Park SK, Pejovic T, Permuth JB, Piskorz A, Prokofyeva D, Riggan MJ, Risch HA, Rodríguez-A… See abstract for full author list ➔ Dareng EO, et al. Am J Hum Genet. 2024 Jun 6;111(6):1061-1083. doi: 10.1016/j.ajhg.2024.04.011. Epub 2024 May 8. Am J Hum Genet. 2024. PMID: 38723632 - Cis- and trans-eQTL TWASs of breast and ovarian cancer identify more than 100 susceptibility genes in the BCAC and OCAC consortia.
Head ST, Dezem F, Todor A, Yang J, Plummer J, Gayther S, Kar S, Schildkraut J, Epstein MP. Head ST, et al. Am J Hum Genet. 2024 Jun 6;111(6):1084-1099. doi: 10.1016/j.ajhg.2024.04.012. Epub 2024 May 8. Am J Hum Genet. 2024. PMID: 38723630 - Exome sequencing identifies HELB as a novel susceptibility gene for non-mucinous, non-high-grade-serous epithelial ovarian cancer.
Dicks EM, Tyrer JP, Ezquina S, Jones M, Baierl J, Peng PC, Diaz M, Goode E, Winham SJ, Dörk T, Van Gorp T, De Fazio A, Bowtell D, Odunsi K, Moysich K, Pavanello M, Campbell I, Brenton JD, Ramus SJ, Gayther SA, Pharoah PDP. Dicks EM, et al. medRxiv [Preprint]. 2024 Apr 3:2024.04.02.24304968. doi: 10.1101/2024.04.02.24304968. medRxiv. 2024. PMID: 38633804 Free PMC article. Preprint. - Large-scale genome-wide association study of 398,238 women unveils seven novel loci associated with high-grade serous epithelial ovarian cancer risk.
Barnes DR, Tyrer JP, Dennis J, Leslie G, Bolla MK, Lush M, Aeilts AM, Aittomäki K, Andrieu N, Andrulis IL, Anton-Culver H, Arason A, Arun BK, Balmaña J, Bandera EV, Barkardottir RB, Berger LPV, de Gonzalez AB, Berthet P, Białkowska K, Bjørge L, Blanco AM, Blok MJ, Bobolis KA, Bogdanova NV, Brenton JD, Butz H, Buys SS, Caligo MA, Campbell I, Castillo C, Claes KBM; GEMO Study Collaborators; EMBRACE Collaborators; Colonna SV, Cook LS, Daly MB, Dansonka-Mieszkowska A, de la Hoya M, deFazio A, DePersia A, Ding YC, Domchek SM, Dörk T, Einbeigi Z, Engel C, Evans DG, Foretova L, Fortner RT, Fostira F, Foti MC, Friedman E, Frone MN, Ganz PA, Gentry-Maharaj A, Glendon G, Godwin AK, González-Neira A, Greene MH, Gronwald J, Guerrieri-Gonzaga A, Hamann U, Hansen TVO, Harris HR, Hauke J, Heitz F, Hogervorst FBL, Hooning MJ, Hopper JL, Huff CD, Huntsman DG, Imyanitov EN; kConFab Investigators; Izatt L, Jakubowska A, James PA, Janavicius R, John EM, Kar S, Karlan BY, Kennedy CJ, Kiemeney LALM, Konstantopoulou I, Kupryjanczyk J, Laitman Y, Lavie O, Lawrenson K, Lester J, Lesueur F, Lopez-Pleguezuelos C, Mai PL, Manoukian S, May T, McNeish IA, Menon U, Milne RL, Modugno F, Mongiovi JM, Montagna M, … See abstract for full author list ➔ Barnes DR, et al. medRxiv [Preprint]. 2024 Mar 4:2024.02.29.24303243. doi: 10.1101/2024.02.29.24303243. medRxiv. 2024. PMID: 38496424 Free PMC article. Preprint. - Phenome-wide association study of ovarian cancer identifies common comorbidities and reveals shared genetics with complex diseases and biomarkers.
Mulugeta A, Lumsden AL, Madakkatel I, Stacey D, Lee SH, Mäenpää J, Oehler MK, Hyppönen E. Mulugeta A, et al. Cancer Med. 2024 Feb;13(4):e7051. doi: 10.1002/cam4.7051. Cancer Med. 2024. PMID: 38457211 Free PMC article.
References
- Pharoah PD, Ponder BA. The genetics of ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2002;16:449–468. - PubMed
- Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA. A systematic review and meta-analysis of family history and risk of ovarian cancer. Br J Obstet Gynaecol. 1998;105:493–499. - PubMed
- Antoniou AC, Easton DF. Risk prediction models for familial breast cancer. Future Oncol. 2006;2:257–274. - PubMed
- Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. - PubMed
Publication types
MeSH terms
Grants and funding
- CA71766/CA/NCI NIH HHS/United States
- 10124/CRUK_/Cancer Research UK/United Kingdom
- R01 CA61107/CA/NCI NIH HHS/United States
- R01 CA058860/CA/NCI NIH HHS/United States
- DH_/Department of Health/United Kingdom
- R01 CA112523-04/CA/NCI NIH HHS/United States
- U01 CA058860/CA/NCI NIH HHS/United States
- R01-CA- 58598/CA/NCI NIH HHS/United States
- 11022/CRUK_/Cancer Research UK/United Kingdom
- CA-58860/CA/NCI NIH HHS/United States
- K07 CA092044-04/CA/NCI NIH HHS/United States
- 11021/CRUK_/Cancer Research UK/United Kingdom
- P30 CA016056/CA/NCI NIH HHS/United States
- R01 CA087538/CA/NCI NIH HHS/United States
- CA16056/CA/NCI NIH HHS/United States
- N01-PC-35137/PC/NCI NIH HHS/United States
- P50 CA105009-05/CA/NCI NIH HHS/United States
- R01 CA87538/CA/NCI NIH HHS/United States
- A6187/CRUK_/Cancer Research UK/United Kingdom
- R01 CA058598/CA/NCI NIH HHS/United States
- N01PC35137/CA/NCI NIH HHS/United States
- K07 CA092044/CA/NCI NIH HHS/United States
- A10124/CRUK_/Cancer Research UK/United Kingdom
- P50 CA105009/CA/NCI NIH HHS/United States
- G0000934/MRC_/Medical Research Council/United Kingdom
- 19275/CRUK_/Cancer Research UK/United Kingdom
- R01 CA122443-02/CA/NCI NIH HHS/United States
- A10119/CRUK_/Cancer Research UK/United Kingdom
- A8339/CRUK_/Cancer Research UK/United Kingdom
- N01-CN-55424/CN/NCI NIH HHS/United States
- R01 CA087538-05/CA/NCI NIH HHS/United States
- R01 CA054419/CA/NCI NIH HHS/United States
- R01-CA-122443/CA/NCI NIH HHS/United States
- R01 CA122443/CA/NCI NIH HHS/United States
- R01 CA114343-03/CA/NCI NIH HHS/United States
- CA-92044/CA/NCI NIH HHS/United States
- 10118/CRUK_/Cancer Research UK/United Kingdom
- R01 CA058598-10/CA/NCI NIH HHS/United States
- R01 CA054419-15/CA/NCI NIH HHS/United States
- R01 CA058860-14/CA/NCI NIH HHS/United States
- P30 CA016056-33/CA/NCI NIH HHS/United States
- R01 CA112523/CA/NCI NIH HHS/United States
- G0801875/MRC_/Medical Research Council/United Kingdom
- P50 CA136393/CA/NCI NIH HHS/United States
- R01 CA114343/CA/NCI NIH HHS/United States
- 10119/CRUK_/Cancer Research UK/United Kingdom
- N01 PC035137/PC/NCI NIH HHS/United States
- R01CA114343/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical