Mining biological pathways using WikiPathways web services - PubMed (original) (raw)
Mining biological pathways using WikiPathways web services
Thomas Kelder et al. PLoS One. 2009.
Abstract
WikiPathways is a platform for creating, updating, and sharing biological pathways [1]. Pathways can be edited and downloaded using the wiki-style website. Here we present a SOAP web service that provides programmatic access to WikiPathways that is complementary to the website. We describe the functionality that this web service offers and discuss several use cases in detail. Exposing WikiPathways through a web service opens up new ways of utilizing pathway information and assisting the community curation process.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
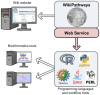
Figure 1. WikiPathways can be accessed by end-users from the wiki-style website.
In addition, the WikiPathways web service provides a programmatic interface that can be used in many programming languages, including R, python, Java and perl and in workflow tools such as Taverna. Using this interface, new pathway analysis tools can be built and existing bioinformatics tools can be extended with pathway-based functionality.
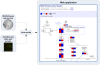
Figure 2. Web application that integrates pathways with gene expression information.
Pathway and gene expression information are retrieved from the WikiPathways and ArrayExpress Atlas web services respectively. A Java servlet integrates this information and publishes it to an interactive web application. In this web application, users can view the information on an interactive pathway diagram.
Similar articles
- Using the Semantic Web for Rapid Integration of WikiPathways with Other Biological Online Data Resources.
Waagmeester A, Kutmon M, Riutta A, Miller R, Willighagen EL, Evelo CT, Pico AR. Waagmeester A, et al. PLoS Comput Biol. 2016 Jun 23;12(6):e1004989. doi: 10.1371/journal.pcbi.1004989. eCollection 2016 Jun. PLoS Comput Biol. 2016. PMID: 27336457 Free PMC article. - WikiPathways: building research communities on biological pathways.
Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. Kelder T, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D1301-7. doi: 10.1093/nar/gkr1074. Epub 2011 Nov 16. Nucleic Acids Res. 2012. PMID: 22096230 Free PMC article. - WikiPathways 2024: next generation pathway database.
Agrawal A, Balcı H, Hanspers K, Coort SL, Martens M, Slenter DN, Ehrhart F, Digles D, Waagmeester A, Wassink I, Abbassi-Daloii T, Lopes EN, Iyer A, Acosta JM, Willighagen LG, Nishida K, Riutta A, Basaric H, Evelo CT, Willighagen EL, Kutmon M, Pico AR. Agrawal A, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D679-D689. doi: 10.1093/nar/gkad960. Nucleic Acids Res. 2024. PMID: 37941138 Free PMC article. - Web tools for predictive toxicology model building.
Jeliazkova N. Jeliazkova N. Expert Opin Drug Metab Toxicol. 2012 Jul;8(7):791-801. doi: 10.1517/17425255.2012.685158. Epub 2012 May 12. Expert Opin Drug Metab Toxicol. 2012. PMID: 22577953 Review. - Review of extracting information from the Social Web for health personalization.
Fernandez-Luque L, Karlsen R, Bonander J. Fernandez-Luque L, et al. J Med Internet Res. 2011 Jan 28;13(1):e15. doi: 10.2196/jmir.1432. J Med Internet Res. 2011. PMID: 21278049 Free PMC article. Review.
Cited by
- A molecular network map of orexin-orexin receptor signaling system.
Chatterjee O, Gopalakrishnan L, Pullimamidi D, Raj C, Yelamanchi S, Gangadharappa BS, Nair B, Mahadevan A, Raju R, Keshava Prasad TS. Chatterjee O, et al. J Cell Commun Signal. 2023 Mar;17(1):217-227. doi: 10.1007/s12079-022-00700-3. Epub 2022 Dec 8. J Cell Commun Signal. 2023. PMID: 36480100 Free PMC article. - Comparison of human cell signaling pathway databases--evolution, drawbacks and challenges.
Chowdhury S, Sarkar RR. Chowdhury S, et al. Database (Oxford). 2015 Jan 28;2015:bau126. doi: 10.1093/database/bau126. Print 2015. Database (Oxford). 2015. PMID: 25632107 Free PMC article. Review. - Computational tools for the interactive exploration of proteomic and structural data.
Morris JH, Meng EC, Ferrin TE. Morris JH, et al. Mol Cell Proteomics. 2010 Aug;9(8):1703-15. doi: 10.1074/mcp.R000007-MCP201. Epub 2010 Jun 4. Mol Cell Proteomics. 2010. PMID: 20525940 Free PMC article. - MetNetAPI: A flexible method to access and manipulate biological network data from MetNet.
Sucaet Y, Wurtele ES. Sucaet Y, et al. BMC Res Notes. 2010 Nov 18;3:312. doi: 10.1186/1756-0500-3-312. BMC Res Notes. 2010. PMID: 21083943 Free PMC article. - ExprEssence--revealing the essence of differential experimental data in the context of an interaction/regulation net-work.
Warsow G, Greber B, Falk SS, Harder C, Siatkowski M, Schordan S, Som A, Endlich N, Schöler H, Repsilber D, Endlich K, Fuellen G. Warsow G, et al. BMC Syst Biol. 2010 Nov 30;4:164. doi: 10.1186/1752-0509-4-164. BMC Syst Biol. 2010. PMID: 21118483 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources