Host responses to intestinal microbial antigens in gluten-sensitive mice - PubMed (original) (raw)
Host responses to intestinal microbial antigens in gluten-sensitive mice
Jane M Natividad et al. PLoS One. 2009.
Abstract
Background and aims: Excessive uptake of commensal bacterial antigens through a permeable intestinal barrier may influence host responses to specific antigen in a genetically predisposed host. The aim of this study was to investigate whether intestinal barrier dysfunction induced by indomethacin treatment affects the host response to intestinal microbiota in gluten-sensitized HLA-DQ8/HCD4 mice.
Methodology/principal findings: HLA-DQ8/HCD4 mice were sensitized with gluten, and gavaged with indomethacin plus gluten. Intestinal permeability was assessed by Ussing chamber; epithelial cell (EC) ultra-structure by electron microscopy; RNA expression of genes coding for junctional proteins by Q-real-time PCR; immune response by in-vitro antigen-specific T-cell proliferation and cytokine analysis by cytometric bead array; intestinal microbiota by fluorescence in situ hybridization and analysis of systemic antibodies against intestinal microbiota by surface staining of live bacteria with serum followed by FACS analysis. Indomethacin led to a more pronounced increase in intestinal permeability in gluten-sensitized mice. These changes were accompanied by severe EC damage, decreased E-cadherin RNA level, elevated IFN-gamma in splenocyte culture supernatant, and production of significant IgM antibody against intestinal microbiota.
Conclusion: Indomethacin potentiates barrier dysfunction and EC injury induced by gluten, affects systemic IFN-gamma production and the host response to intestinal microbiota antigens in HLA-DQ8/HCD4 mice. The results suggest that environmental factors that alter the intestinal barrier may predispose individuals to an increased susceptibility to gluten through a bystander immune activation to intestinal microbiota.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
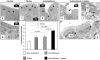
Figure 1. Intestinal barrier measurements.
Ussing-chamber experiments were performed on jejunum from all four groups 24 hours after the last gluten challenge. (A) Gluten-sensitized mice treated with indomethacin showed a significant increase in tissue conductance. (B) HRP flux (transcellular permeability) increased significantly in all treatment groups compared to non-sensitized controls, however the highest values were observed in gluten plus indomethacin treated mice. Data represent the means±SEM of 10 mice/group.
Figure 2. Evaluation of mitochondrial disruption.
Mitochondrial ultra-structure was assessed by electron microscopy. Indomethacin increased the fraction of disrupted mitochondria. Gluten sensitization plus indomethacin treatment further increased the proportion of altered mitochondria. Data represent the means±SEM of 6 mice/group. Reperesentative pictures from (A) Control mouse, arrow: normal mitochondria; (B) Gluten-sensitized mouse; (C) Indomethacin-treated mouse, arrowhead: mitochondria with disrupted cristae; (D) Indomethacin-treated plus gluten-sensitized mouse, arrowhead: mitochondria with disrupted cristae.
Figure 3. Apical epithelial cell structural abnormalities.
Epithelial ultra-structure was assessed by electron microscopy. A significant proportion of altered TJ was observed in gluten-sensitized plus indomethacin-treated mice. Indomethacin alone also increased the proportion of altered TJ but to a lesser extent than indomethacin plus gluten. Gluten sensitization alone tended to increase the proportion of altered TJ but this did not achieve statistical significance (p = 0.09 vs non-sensitized controls). Data represent the means±SEM of 5 mice/group. Representative pictures of (A–B) a control mouse, arrow: tight junction (TJ) with preserved structure; (C) Gluten-sensitized mouse, arrow: TJ with preserved structure; (D) Indomethacin-treated mouse showing one altered TJ (arrowhead) and 2 junctions with normal struture (arrows); (E–G) Gluten plus indometacin treated mouse; (E) arrowhead: microvilli (mv) height reduction, arrow: apical epithelial cell destruction; (F) Altered TJ, arrow: mitochondria (m) with disrupted cristae; (G) Several altered TJs.
Figure 4. RNA level of E-cadherin relative to control (non-sensitized).
Real-time QPCR experiments were performed on jejunum collected from all groups 24 hours after the last gluten challenge. Gluten-sensitized and indomethacin alone-treated mice showed a trend for decreased expression of E-cadherin relative to non-sensitized controls. Gluten-sensitized plus indomethacin-treated mice showed marked down-regulation of E-cadherin RNA level relative to non-sensitized controls. Data represent the means±SEM of 6 mice/group.
Figure 5. Splenocyte proliferation after incubation with PT gliadin.
Proliferation was measured by 3H-thymidine incorporation and expressed as stimulation index. Splenocytes from gluten-sensitized mice treated with or without indomethacin exhibited increased proliferation compared to non-sensitized controls. Data represent the means±SEM of 6 mice/group.
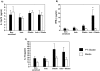
Figure 6. Cytokines in supernatant of splenocyte cultures after incubation with PT-gliadin (black) or medium (white).
Expressions of (A) IL-12p70, (B) IFN-γ, (C) IL-10 were determined by CBA analysis. Culture supernatants from gluten-sensitized plus indomethacin (Indo) treated mice showed increased IFN- γ (*p<0.01 vs all groups). Cultured splenocytes from gluten-sensitized mice, with or without indomethacin showed a trend for increased IL-10 release after PT-gliadin stimulation (p = 0.09). Data represent the means±SEM of 6 mice/group.
Figure 7. Microbiota composition.
Using 9 different oligonucleotide probes and fluorescent in situ hybridization (FISH), microbial profile was investigated in the distal jejunum of sensitized mice with and without indomethacin. The results indicate a significant perturbation in the proportions of microbiota investigated in all 3 treatment-groups when compared to non-sensitized controls, and remarkably in the gluten-sensitized plus indomethacin group. These differences achieve statistical significance in Bifidobacteria (*p = 0.04 vs controls, +p = 0.03 vs gluten) and Clostridium Leptum cluster (both *p = 0.02 vs controls and gluten sensitized, **p = 0.04 vs indomethacin) compared to gluten-sensitized alone. Data represent the means±SEM of 6 mice/group.
Figure 8. Systemic antibodies against commensals.
Serum from indomethacin and gluten treated mice showed significant positive serum antibodies against their aerobic and anaerobic intestinal microbiota. (A) Representative FACS histogram from each treatment gated on IgM+ cells; (B) Proportion of IgM+ aerobic bacterial cells for each treatment groups; (C) Proportion of IgM+ anaerobic bacterial cells for each treatment groups. Data represent the means±SEM of 6 mice/group.
Similar articles
- HLA-DQ determines the response to exogenous wheat proteins: a model of gluten sensitivity in transgenic knockout mice.
Black KE, Murray JA, David CS. Black KE, et al. J Immunol. 2002 Nov 15;169(10):5595-600. doi: 10.4049/jimmunol.169.10.5595. J Immunol. 2002. PMID: 12421937 - Gliadin-dependent neuromuscular and epithelial secretory responses in gluten-sensitive HLA-DQ8 transgenic mice.
Verdu EF, Huang X, Natividad J, Lu J, Blennerhassett PA, David CS, McKay DM, Murray JA. Verdu EF, et al. Am J Physiol Gastrointest Liver Physiol. 2008 Jan;294(1):G217-25. doi: 10.1152/ajpgi.00225.2007. Epub 2007 Nov 15. Am J Physiol Gastrointest Liver Physiol. 2008. PMID: 18006603 - Increased bacterial translocation in gluten-sensitive mice is independent of small intestinal paracellular permeability defect.
Silva MA, Jury J, Sanz Y, Wiepjes M, Huang X, Murray JA, David CS, Fasano A, Verdú EF. Silva MA, et al. Dig Dis Sci. 2012 Jan;57(1):38-47. doi: 10.1007/s10620-011-1847-z. Epub 2011 Aug 7. Dig Dis Sci. 2012. PMID: 21822909 Free PMC article. - Microbiome and Gluten.
Sanz Y. Sanz Y. Ann Nutr Metab. 2015;67 Suppl 2:28-41. doi: 10.1159/000440991. Epub 2015 Nov 26. Ann Nutr Metab. 2015. PMID: 26605783 Review. - Celiac disease: Autoimmunity in response to food antigen.
Stamnaes J, Sollid LM. Stamnaes J, et al. Semin Immunol. 2015 Sep;27(5):343-52. doi: 10.1016/j.smim.2015.11.001. Epub 2015 Nov 18. Semin Immunol. 2015. PMID: 26603490 Review.
Cited by
- The Infant-Derived Bifidobacterium bifidum Strain CNCM I-4319 Strengthens Gut Functionality.
Martín R, Bottacini F, Egan M, Chamignon C, Tondereau V, Moriez R, Knol J, Langella P, Eutamene H, Smokvina T, van Sinderen D. Martín R, et al. Microorganisms. 2020 Aug 28;8(9):1313. doi: 10.3390/microorganisms8091313. Microorganisms. 2020. PMID: 32872165 Free PMC article. - Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice.
Laval L, Martin R, Natividad JN, Chain F, Miquel S, Desclée de Maredsous C, Capronnier S, Sokol H, Verdu EF, van Hylckama Vlieg JE, Bermúdez-Humarán LG, Smokvina T, Langella P. Laval L, et al. Gut Microbes. 2015;6(1):1-9. doi: 10.4161/19490976.2014.990784. Epub 2015 Jan 14. Gut Microbes. 2015. PMID: 25517879 Free PMC article. - Latest in vitro and in vivo models of celiac disease.
Stoven S, Murray JA, Marietta EV. Stoven S, et al. Expert Opin Drug Discov. 2013 Apr;8(4):445-57. doi: 10.1517/17460441.2013.761203. Epub 2013 Jan 8. Expert Opin Drug Discov. 2013. PMID: 23293929 Free PMC article. Review. - Animal models to study gluten sensitivity.
Marietta EV, Murray JA. Marietta EV, et al. Semin Immunopathol. 2012 Jul;34(4):497-511. doi: 10.1007/s00281-012-0315-y. Epub 2012 May 11. Semin Immunopathol. 2012. PMID: 22572887 Free PMC article. Review. - The incidence and risk of celiac disease in a healthy US adult population.
Riddle MS, Murray JA, Porter CK. Riddle MS, et al. Am J Gastroenterol. 2012 Aug;107(8):1248-55. doi: 10.1038/ajg.2012.130. Epub 2012 May 15. Am J Gastroenterol. 2012. PMID: 22584218 Free PMC article.
References
- Cronin CC, Shanahan F. Exploring the iceberg-the spectrum of celiac disease. Am J Gastroenterol. 2003;98:518–520. - PubMed
- Rostom A, Murray JA, Kagnoff MF. American Gastroenterological Association (AGA) Institute technical review on the diagnosis and management of celiac disease. Gastroenterology. 2006;131:1981–2002. - PubMed
- Troncone R, Franzese A, Mazzarella G, Paparo F, Auricchio R, et al. Gluten sensitivity in a subset of children with insulin dependent diabetes mellitus. Am J Gastroenterol. 2003;98:590–595. - PubMed
- Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology. 1992;102:330–54. - PubMed
- Kaukinen K, Maki M, Partanen J, Sievanen H, Collin P. Celiac disease without villous atrophy: revision of criteria called for. Dig Dis Sci. 2001;46:879–887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials