A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction - PubMed (original) (raw)
A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction
Felipe Diaz-Griffero et al. J Virol. 2009 Oct.
Abstract
TRIM5alpha is a tripartite motif (TRIM) protein that consists of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains. The TRIM5alpha(rh) protein from rhesus monkeys recognizes the human immunodeficiency virus type 1 (HIV-1) capsid as it enters the host cell and blocks virus infection prior to reverse transcription. HIV-1-restricting ability can be eliminated by disruption of the B-box 2 domain. Changes in the TRIM5alpha(rh) B-box 2 domain have been associated with alterations in TRIM5alpha(rh) turnover, the formation of cytoplasmic bodies and higher-order oligomerization. We present here the nuclear magnetic resonance structure of the TRIM5 B-box 2 domain and identify an unusual hydrophobic patch (cluster 1) on the domain surface. Alteration of cluster 1 or the flanking arginine 121 resulted in various degrees of inactivation of HIV-1 restriction, in some cases depending on compensatory changes in other nearby charged residues. For this panel of TRIM5alpha(rh) B-box 2 mutants, inhibition of HIV-1 infection was strongly correlated with higher-order self-association and binding affinity for capsid complexes but not with TRIM5alpha(rh) half-life or the formation of cytoplasmic bodies. Thus, promoting cooperative TRIM5alpha(rh) interactions with the HIV-1 capsid represents a major mechanism whereby the B-box 2 domain potentiates HIV-1 restriction.
Figures
FIG. 1.
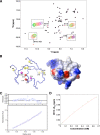
Solution structure of the human TRIM5 B-box 2 domain. (A) Superposition of 20 NMR structures showing the α-helix in red, the β-strands in blue, and the Zn2+ in green. (B) Ribbon diagram of the NMR structure shown in panel A from the same perspective. The Zn2+ coordinating residues (CHCDC2H2) are also shown in green. (C) Electrostatic mapping of the B-box 2 surface, highlighting the positions of the positive (blue), negative (red), or neutral (white) charges.
FIG. 2.
B-box 2 domains of human and rhesus monkey TRIM5. (A) The human and rhesus monkey TRIM5 B-box 2 domain sequences are aligned and numbered. Residues with surface-accessible side chains are colored as follows: green, hydrophobic; red, acidic; and blue, basic. (B) A Corey-Pauling-Koltun model of the human and rhesus monkey TRIM5 B-box 2 domain is shown. The residue numbers correspond to those of human TRIM5, with the residue numbers of rhesus monkey TRIM5 in parentheses. The orientations of the left and right figures are the same as those of the upper and lower figures, respectively, in Fig. 1; the right image represents a rotation of the left image 180° around the vertical axis. Acidic and basic residues are shown in red and blue, respectively. Hydrophobic residues are shown in green. Cluster 1 is evident in the left figure, and cluster 2 is evident in the right figure.
FIG. 3.
HSQC perturbation analysis in concentration-dependent experiments. (A) Eleven 1H-15N HSQC spectra were measured at protein concentrations of 660, 560, 490, 400, 300, 250, 170, 110, 40, 20, and 10 μM and superimposed, with colors ranging from purple (highest concentration) to green (lowest concentration). Signal appearances were observed for the backbone amide proton of E118 and imino proton of the W115 indole ring at the lower concentrations. Chemical shift changes and sharpening were observed for other signals. (B) Mapping of perturbed residues on the B-box 2 structure. In the figure on the left, the size of the CSP is indicated by color. The red color means more than 0.03 ppm of CSP were observed, and yellow indicates more than 0.01 ppm of CSP. The magenta color indicates the appearance of the signal at a lower concentration. Most of the implicated residues are proximal to cluster 1 on one face of the B-box 2 domain (dotted circle on the electrostatic surface on the right). (C) Equilibrium analytical ultracentrifugation of the TRIM5 B-box 2 domain. The radial distribution of the absorbance in the centrifuge cell at equilibrium at 22,000 rpm, with a protein concentration of 1.2 mg/ml (∼180 μM), is shown in the lower panel. The red line through the data represents the fit to a single species with a molecular weight corresponding to 10,502; the theoretical molecular weight for the single polypeptide chain is 6,682. The upper panel shows the residuals for the fit. (D) The concentration dependence of chemical shift perturbation for the W115 backbone amide proton in 1H-15N HSQC spectra. Total chemical shift change was calculated by using the relation: Δδtot = [(δHN)2 + (δN/6.5)2]1/2 (1). Curve fitting was carried out by using the program gnuplot. The calculated equilibrium dissociation constant KD was 9.33 ± 5.40 mM (see the text).
FIG. 4.
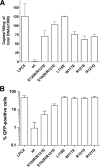
Restriction of HIV-1 infection by TRIM5αrh mutants. Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. Control cells were transduced with the empty LPCX vector. Stably expressing cell lines were selected with 5 μg of puromycin/ml. The cells were incubated with HIV-1-GFP, and the percentage of GFP-positive cells was measured 48 h later by FACS. The results of two independent experiments were similar; the results of a single experiment are shown.
FIG. 5.
Blockade of HIV-1 reverse transcription by the TRIM5αrh B-box 2 mutants. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αrh proteins or containing the empty LPCX vector were challenged at a multiplicity of infection of 0.4 with DNase-pretreated HIV-1-GFP. After 6 h, the cells were lysed, and the total DNA was extracted. (A) The levels of viral DNA were measured by quantitative real-time PCR, using a probe against GFP, as described in Materials and Methods. The means and standard deviations of the results derived from three independent experiments performed in triplicate are shown. (B) In some cases, infections were left to proceed for 48 h, and infectivity was measured as the percentage of GFP-positive cells by FACS. The values shown represent the means and standard deviations of the results obtained in three independent experiments.
FIG. 6.
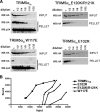
Binding of TRIM5αrh B-box 2 mutants to assembled HIV-1 capsids. (A) 293T cells were transfected with plasmids expressing the indicated wild-type and mutant TRIM5αrh proteins tagged with HA epitopes. At 36 h after transfection, the cells were lysed. Serial dilutions of the cleared lysates were made by mixing with cleared lysates from untransfected 293T cells. The lysates were incubated at room temperature for 1 h with HIV-1 CA-NC complexes that had been assembled in vitro. The mixtures were applied to a 70% sucrose cushion and centrifuged. Input represents the mixtures analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted for the HA tag. The pellet from the 70% cushion (pellet) was analyzed by Western blotting with antibodies against the HA tag and HIV-1 CA-NC protein. (B) The Western blots shown in panel A were quantitated as described in Materials and Methods. The amounts of TRIM5αrh protein in the input and pellet (bound) fractions are shown in arbitrary units.
FIG. 7.
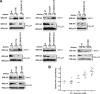
Binding of additional TRIM5αrh mutants to assembled HIV-1 capsid complexes. (A) The binding of the indicated TRIM5αrh variants to HIV-1 CA-NC complexes assembled in vitro were assessed as described in the Fig. 6 legend and Materials and Methods. (B) The relative capsid-binding ability and HIV-1-restricting activity of each TRIM5αrh B-box 2 variant were assessed as described in the Table 2 footnotes and in Materials and Methods. The means for the relative capsid-binding abilities associated with each HIV-1 restriction group are indicated by horizontal bars. The Spearman rank correlation coefficient, _r_s, is 0.8421, with a 95% confidence interval of 0.779 to 0.905 (two-sided P value of <0.001).
FIG. 8.
Higher-order self-association of TRIM5αrh B-box 2 mutants. 293T cells were transfected with plasmids expressing the indicated wild-type or mutant TRIM5αrh proteins with a FLAG or an HA epitope tag. Cells expressing wild-type and mutant TRIM5αrh proteins were lysed 48 h after transfection. (A to D) The cell lysates were mixed, and the indicated mixtures were used for immunoprecipitation by an antibody directed against the FLAG epitope, as described in Materials and Methods. Elution of the immunocomplexes was performed with a FLAG tripeptide and analyzed by Western blotting with anti-HA and anti-FLAG antibodies. WB, Western blot; IP, immunoprecipitation. (E) The higher-order self-association and HIV-1-restricting ability, determined as described in the Table 2 footnotes and in Materials and Methods, of each TRIM5αrh variant were compared. The means of the values for higher-order self-association within each restriction group are indicated by horizontal bars. The Spearman rank correlation coefficient, _r_s, is 0.862, with a 95% confidence interval of 0.665 to 1.0 (two-sided P value of <0.001).
FIG. 9.
Model for the role of the TRIM5α B-box 2 domain in HIV-1 restriction. The TRIM5αrh dimer is depicted, with the B-box 2 domains (magenta) and B30.2(SPRY) domains (blue) highlighted. Interactions involving the cluster 1 region of the B-box 2 domain increase the avidity of TRIM5αrh for the retroviral capsid, promoting the aggregation of the TRIM5αrh protein on the capsid surface.
Similar articles
- General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.
Wagner JM, Christensen DE, Bhattacharya A, Dawidziak DM, Roganowicz MD, Wan Y, Pumroy RA, Demeler B, Ivanov DN, Ganser-Pornillos BK, Sundquist WI, Pornillos O. Wagner JM, et al. J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article. - The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association.
Li X, Sodroski J. Li X, et al. J Virol. 2008 Dec;82(23):11495-502. doi: 10.1128/JVI.01548-08. Epub 2008 Sep 17. J Virol. 2008. PMID: 18799578 Free PMC article. - Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains.
Li X, Li Y, Stremlau M, Yuan W, Song B, Perron M, Sodroski J. Li X, et al. J Virol. 2006 Jul;80(13):6198-206. doi: 10.1128/JVI.00283-06. J Virol. 2006. PMID: 16775307 Free PMC article. - Anti-retroviral activity of TRIM5 alpha.
Nakayama EE, Shioda T. Nakayama EE, et al. Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review. - Retroviral restriction factors TRIM5α: therapeutic strategy to inhibit HIV-1 replication.
Zhang J, Ge W, Zhan P, De Clercq E, Liu X. Zhang J, et al. Curr Med Chem. 2011;18(17):2649-54. doi: 10.2174/092986711795933687. Curr Med Chem. 2011. PMID: 21568899 Review.
Cited by
- A novel aminoacid determinant of HIV-1 restriction in the TRIM5α variable 1 region isolated in a random mutagenic screen.
Pham QT, Veillette M, Brandariz-Nuñez A, Pawlica P, Thibert-Lefebvre C, Chandonnet N, Diaz-Griffero F, Berthoux L. Pham QT, et al. Virus Res. 2013 May;173(2):306-314. doi: 10.1016/j.virusres.2013.01.013. Epub 2013 Jan 25. Virus Res. 2013. PMID: 23357295 Free PMC article. - Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.
Sanchez JG, Chiang JJ, Sparrer KMJ, Alam SL, Chi M, Roganowicz MD, Sankaran B, Gack MU, Pornillos O. Sanchez JG, et al. Cell Rep. 2016 Aug 2;16(5):1315-1325. doi: 10.1016/j.celrep.2016.06.070. Epub 2016 Jul 14. Cell Rep. 2016. PMID: 27425606 Free PMC article. - Adaptation of HIV-1 to cells expressing rhesus monkey TRIM5α.
Pacheco B, Finzi A, Stremlau M, Sodroski J. Pacheco B, et al. Virology. 2010 Dec 20;408(2):204-12. doi: 10.1016/j.virol.2010.09.019. Epub 2010 Oct 16. Virology. 2010. PMID: 20956011 Free PMC article. - Unique spectrum of activity of prosimian TRIM5alpha against exogenous and endogenous retroviruses.
Rahm N, Yap M, Snoeck J, Zoete V, Muñoz M, Radespiel U, Zimmermann E, Michielin O, Stoye JP, Ciuffi A, Telenti A. Rahm N, et al. J Virol. 2011 May;85(9):4173-83. doi: 10.1128/JVI.00075-11. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345948 Free PMC article. - Modulation of TRIM5alpha activity in human cells by alternatively spliced TRIM5 isoforms.
Battivelli E, Migraine J, Lecossier D, Matsuoka S, Perez-Bercoff D, Saragosti S, Clavel F, Hance AJ. Battivelli E, et al. J Virol. 2011 Aug;85(15):7828-35. doi: 10.1128/JVI.00648-11. Epub 2011 Jun 1. J Virol. 2011. PMID: 21632761 Free PMC article.
References
- Ayed, A., F. A. Mulder, G. S. Yi, Y. Lu, L. E. Kay, and C. H. Arrowsmith. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8:756-760. - PubMed
- Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. - PubMed
- Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. - PubMed
- Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI063987/AI/NIAID NIH HHS/United States
- R21 AI076094/AI/NIAID NIH HHS/United States
- R00 MH086162/MH/NIMH NIH HHS/United States
- AI60354/AI/NIAID NIH HHS/United States
- AI063987/AI/NIAID NIH HHS/United States
- 1K99MH086162-01/MH/NIMH NIH HHS/United States
- K99 MH086162/MH/NIMH NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
- AI076094/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources