Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia - PubMed (original) (raw)
Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia
Jennifer L Umbach et al. J Virol. 2009 Oct.
Abstract
Analysis of cells infected by a wide range of herpesviruses has identified numerous virally encoded microRNAs (miRNAs), and several reports suggest that these viral miRNAs are likely to play key roles in several aspects of the herpesvirus life cycle. Here we report the first analysis of human ganglia for the presence of virally encoded miRNAs. Deep sequencing of human trigeminal ganglia latently infected with two pathogenic alphaherpesviruses, herpes simplex virus 1 (HSV-1) and varicella-zoster virus (VZV), confirmed the expression of five HSV-1 miRNAs, miR-H2 through miR-H6, which had previously been observed in mice latently infected with HSV-1. In addition, two novel HSV-1 miRNAs, termed miR-H7 and miR-H8, were also identified. Like four of the previously reported HSV-1 miRNAs, miR-H7 and miR-H8 are encoded within the second exon of the HSV-1 latency-associated transcript. Although VZV genomic DNA was readily detectable in the three human trigeminal ganglia analyzed, we failed to detect any VZV miRNAs, suggesting that VZV, unlike other herpesviruses examined so far, may not express viral miRNAs in latently infected cells.
Figures
FIG. 1.
Two novel HSV-1 miRNAs recovered from latently infected humans. (A) Predicted secondary structures of the primary miRNA stem-loops of the novel HSV-1 miRNAs miR-H7 and miR-H8. Arrowheads indicate sites of Drosha cleavage, and arrows indicate sites of Dicer cleavage. The recovered, mature 5p and 3p miRNA strands are shown in boldface. (B) Schematic of the HSV-1 genome expanded to show details of the LAT locus. Relative sizes, locations, and orientations of other viral transcripts are also indicated. Sequence coordinates of viral miRNAs are given according to the HSV-1 strain 17 syn+ genome (NC_001806). IR, internal repeat; TR, terminal repeat; UL, unique long; US, unique short. The novel HSV-1 miRNAs miR-H7 and miR-H8 both map antisense to an intronic region in the viral ICP0 gene.
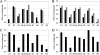
FIG. 2.
HSV-1 miRNA expression determined by qRT-PCR in latently and productively infected cells and tissues. All values are given as changes in expression relative to that of a non-HSV-1-infected control sample and were normalized to values for a cellular miRNA, miR-16. (A) HSV-1 miRNA expression levels in latently infected human TG from subject 1 (filled bars) and subject 2 (shaded bars). Despite its recovery during deep sequencing, miR-H8 was not detected in either TG sample. Subject 3 served as the negative control. (B) HSV-1 miRNA expression profiles, at 6 hpi (solid bars) and 18 hpi (shaded bars), of SY5Y cells productively infected with HSV-1 strain KOS at a multiplicity of infection of 10. (C and D) Similar to panel B, except that results for HeLa cells at 18 hpi are shown. (D) Similar to panel B, except that Vero cells at 24 hpi were analyzed.
Similar articles
- Herpes simplex virus 1 miRNA sequence variations in latently infected human trigeminal ganglia.
Cokarić Brdovčak M, Zubković A, Ferenčić A, Šoša I, Stemberga V, Cuculić D, Rokić F, Vugrek O, Hackenberg M, Jurak I. Cokarić Brdovčak M, et al. Virus Res. 2018 Sep 2;256:90-95. doi: 10.1016/j.virusres.2018.08.002. Epub 2018 Aug 2. Virus Res. 2018. PMID: 30077725 - Mutations Inactivating Herpes Simplex Virus 1 MicroRNA miR-H2 Do Not Detectably Increase ICP0 Gene Expression in Infected Cultured Cells or Mouse Trigeminal Ganglia.
Pan D, Pesola JM, Li G, McCarron S, Coen DM. Pan D, et al. J Virol. 2017 Jan 3;91(2):e02001-16. doi: 10.1128/JVI.02001-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847363 Free PMC article. - MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs.
Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. Umbach JL, et al. Nature. 2008 Aug 7;454(7205):780-3. doi: 10.1038/nature07103. Epub 2008 Jul 2. Nature. 2008. PMID: 18596690 Free PMC article. - A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.
Kennedy PG, Rovnak J, Badani H, Cohrs RJ. Kennedy PG, et al. J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review. - Mammalian alphaherpesvirus miRNAs.
Jurak I, Griffiths A, Coen DM. Jurak I, et al. Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):641-53. doi: 10.1016/j.bbagrm.2011.06.010. Epub 2011 Jun 28. Biochim Biophys Acta. 2011. PMID: 21736960 Free PMC article. Review.
Cited by
- MicroRNAs as mediators of viral evasion of the immune system.
Cullen BR. Cullen BR. Nat Immunol. 2013 Mar;14(3):205-10. doi: 10.1038/ni.2537. Epub 2013 Feb 15. Nat Immunol. 2013. PMID: 23416678 Free PMC article. Review. - Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture.
Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. Flores O, et al. J Virol. 2013 Jun;87(12):6589-603. doi: 10.1128/JVI.00504-13. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536669 Free PMC article. - The molecular basis of herpes simplex virus latency.
Nicoll MP, Proença JT, Efstathiou S. Nicoll MP, et al. FEMS Microbiol Rev. 2012 May;36(3):684-705. doi: 10.1111/j.1574-6976.2011.00320.x. Epub 2012 Jan 10. FEMS Microbiol Rev. 2012. PMID: 22150699 Free PMC article. Review. - MicroRNAs: Harbingers and shapers of periodontal inflammation.
Luan X, Zhou X, Fallah P, Pandya M, Lyu H, Foyle D, Burch D, Diekwisch TGH. Luan X, et al. Semin Cell Dev Biol. 2022 Apr;124:85-98. doi: 10.1016/j.semcdb.2021.05.030. Epub 2021 Jun 10. Semin Cell Dev Biol. 2022. PMID: 34120836 Free PMC article. Review. - Viral miRNAs: tools for immune evasion.
Boss IW, Renne R. Boss IW, et al. Curr Opin Microbiol. 2010 Aug;13(4):540-5. doi: 10.1016/j.mib.2010.05.017. Epub 2010 Jun 25. Curr Opin Microbiol. 2010. PMID: 20580307 Free PMC article. Review.
References
- Bloom, D. C. 2004. HSV LAT and neuronal survival. Int. Rev. Immunol. 23:187-198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI067968/AI/NIAID NIH HHS/United States
- R01 AI067968/AI/NIAID NIH HHS/United States
- AI067968/AI/NIAID NIH HHS/United States
- P01 AG032958/AG/NIA NIH HHS/United States
- T32 CA009111/CA/NCI NIH HHS/United States
- AG032958/AG/NIA NIH HHS/United States
- T32 NS007321/NS/NINDS NIH HHS/United States
- R01 AI067968-04/AI/NIAID NIH HHS/United States
- R01 AG006127/AG/NIA NIH HHS/United States
- AG006127/AG/NIA NIH HHS/United States
- T32-CA009111/CA/NCI NIH HHS/United States
- R37 AG006127/AG/NIA NIH HHS/United States
- NS007321/NS/NINDS NIH HHS/United States
- T32 CA009111-32/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources