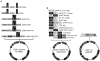A versatile viral system for expression and depletion of proteins in mammalian cells - PubMed (original) (raw)
A versatile viral system for expression and depletion of proteins in mammalian cells
Eric Campeau et al. PLoS One. 2009.
Abstract
The ability to express or deplete proteins in living cells is crucial for the study of biological processes. Viral vectors are often useful to deliver DNA constructs to cells that are difficult to transfect by other methods. Lentiviruses have the additional advantage of being able to integrate into the genomes of non-dividing mammalian cells. However, existing viral expression systems generally require different vector backbones for expression of cDNA, small hairpin RNA (shRNA) or microRNA (miRNA) and provide limited drug selection markers. Furthermore, viral backbones are often recombinogenic in bacteria, complicating the generation and maintenance of desired clones. Here, we describe a collection of 59 vectors that comprise an integrated system for constitutive or inducible expression of cDNAs, shRNAs or miRNAs, and use a wide variety of drug selection markers. These vectors are based on the Gateway technology (Invitrogen) whereby the cDNA, shRNA or miRNA of interest is cloned into an Entry vector and then recombined into a Destination vector that carries the chosen viral backbone and drug selection marker. This recombination reaction generates the desired product with >95% efficiency and greatly reduces the frequency of unwanted recombination in bacteria. We generated Destination vectors for the production of both retroviruses and lentiviruses. Further, we characterized each vector for its viral titer production as well as its efficiency in expressing or depleting proteins of interest. We also generated multiple types of vectors for the production of fusion proteins and confirmed expression of each. We demonstrated the utility of these vectors in a variety of functional studies. First, we show that the FKBP12 Destabilization Domain system can be used to either express or deplete the protein of interest in mitotically-arrested cells. Also, we generate primary fibroblasts that can be induced to senesce in the presence or absence of DNA damage. Finally, we determined that both isoforms of the AT-Rich Interacting Domain 4B (ARID4B) protein could induce G1 arrest when overexpressed. As new technologies emerge, the vectors in this collection can be easily modified and adapted without the need for extensive recloning.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
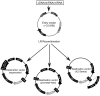
Figure 1. Overview of the viral system.
A cDNA/shRNA/miRNA is cloned into an Entry vector between the attL1 and attL2 sites. In the presence of the LR clonase, recombination occurs between attL1-attR1 and attL2-attR2 to transfer the insert from the Entry vector into the Destination vector of choice. All Entry vectors contain the kanamycin resistance gene whereas all Destination vectors carry the ampicillin resistance gene.
Figure 2. Maps of the Entry vectors for either RNAi-mediated protein depletion or protein overexpression.
A) shRNAs or miRNAs can be expressed with either constitutive (H1, U6, CMV) or inducible (H1/TO, CMV/TO) promoters. B) cDNAs encoding the protein of interest (POI) can be cloned into Entry vectors with no tags or different tags for either detection or purification. (Abbreviations: CBP, calcium-binding peptide; SBP, S-binding peptide; GST, glutathione-S-transferase; FKBP12DD, FKBP destabilizing domain. LEFT: Promoter-less Entry vectors. RIGHT: Entry vectors for EF-1α promoter-driven protein expression. In this case, Entry vectors containing a promoter should be recombined with a promoter-less Destination vector (see Figure 4).
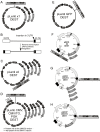
Figure 3. Maps of the Destination vectors.
(A, C) The pLenti X1 and X2 series are promoter-less and require that the expression promoter come from the Entry vector. (B) The pLenti X2 series is designed for shRNA insertion into the 3′ LTR, resulting in insert duplication in the final integrated form of the viral genome. (D) The CMV and PGK series provide the promoters for constitutive expression, and the CMV/TO promoter for regulation of expression by doxycycline. (E) The pLenti GFP DEST allows insertion of an expression cassette after the Woodchuck post-transcriptional element (PRE) and expresses GFP to make recipient cells fluorescent. (F) Retroviral Destination vectors for RNAi where the Destination cassette was inserted in the 3′ LTR, similar to the pLenti X2 series. (G, H). Retroviral Destination vectors for cDNA expression under the control of the CMV promoter. See text, Table 1 and Figure 4 for details and compatibilities between each vector.
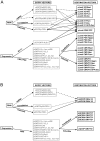
Figure 4. Overview of the lentiviral (A) or retroviral (B) vectors for protein expression or depletion.
Entry vectors can be recombined using the LR clonase into the Destination vectors connected by arrows. For RNAi-mediated depletion, miRNA-based and shRNA-based vectors are available for either constitutive or inducible depletions (see Figure 2A for details). For expression, various tags are available (see Figure 2B for details). The RNAi cassettes can be inserted in the middle of the lentiviral backbone (pLenti X1 series), in the 3′ LTR (pLenti X2 series) or in a backbone expressing GFP (pLenti CMV GFP). For RNAi studies using retroviruses, all the Destination cassettes have been inserted in the 3′ LTR of the vectors. For protein expression, the EF-1α, PGK or CMV promoters are available for lentiviruses. CMV-driven constructs can be either constitutive or inducible. For the retroviral vectors, no constitutive expression of cDNAs can be attained in T-REx cell lines using the pQCXI series vectors because they all contain the inducible CMV/TO promoter. Similarly, the drug resistance gene will be repressed in a T-REx cell line since it is after an IRES element under the control of the CMV/TO promoter. However, the pQCXP CMV/TO can be used for inducible expression of cDNAs under constitutive puromycin selection in T-REx cell lines. See Table 2 for details.
Figure 5. Comparison of titer efficiencies of various lentiviral vectors available to the scientific community.
Twenty-five thousand HeLa cells were seeded in each well of a six-well plate and transduced with either 10, 1 or 0.1 µl of viral supernatants. Forty-eight hours post-transduction, puromycin was added at a final concentration of 0.5 µg/ml. Twelve days post-transduction, the cells were fixed and stained with crystal violet. The LKO.1, GIPZ and our viral vector (pLenti CMV/TO) yielded similar titers (2–8×105 cfu/ml) whereas the FSIPPW yielded a higher titer (∼7×106 cfu/ml).
Figure 6. Depletion of Asf1a and MDC1 using various lentiviral vectors.
A) Asf1a depletion in U2OS cells. Lane 1: U2OS T-REx cell extracts, untransduced. Lane 2 and 3: U2OS transduced with two different miRNAs against Asf1a in the pLenti X2 Hygro/pSM2(CMV) vector; Lane 4 and 5: U2OS T-REx cells transduced with the pLenti X2 Neo/pTER shAsf1a #1 plasmid either uninduced (lane 4) or induced for 96 h (lane 5). γ-tubulin levels were monitored as a loading control for protein levels. (B) Depletion of MDC1 using the pLenti X1 GFP-Zeo vector. TOP: Map of recombinant lentivirus used to deplete MDC1 with the GFP-Zeocin fusion protein. BOTTOM: Immunofluorescent detection of MDC1 knockdown. Transduced cells produce the GFP-Zeocin resistance fusion protein, and therefore can be detected by green fluorescence (left panel). MDC1 protein was detected using an MDC1 antibody and an Alexa568-labeled secondary antibody, shown in the red channel (middle panel). Merging of these two images (right panel) indicates that the cells expressing the GFP fusion protein are depleted of MDC1. (C) Inducible depletion of MDC1 in BJ T-REx fibroblasts using the pLenti X1 Zeo vector. The cells were induced for 96 h and collected for Western blot analysis. The same MDC1 antibody used in (B) was used for the detection of MDC1. A cross-reacting band (indicated by the asterisk, *) is used as a loading control. It is not depleted by the MDC1 shRNA and therefore, appears to be an endogenous cellular protein that is recognized fortuitously on immunoblots.
Figure 7. Use of the lentiviral Destination vectors to overexpress proteins.
(A) Generation of three VA13 cell lines inducibly expressing either V5-HIRA, V5-HIRA and HA-Asf1a, or V5-HIRA and HA-Asf1b. (A) Cells were first transduced with the pLenti TetR Blast lentivirus and selected with blasticidin. These cells where then transduced with either the pLenti CMV/TO V5-HIRA Hygro virus alone or in combination with either the pLenti CMV/TO HA-Asf1a Puro or pLenti CMV/TO HA-Asf1b Puro virus. After appropriate drug selection, the cells were tested for the expression of the respective proteins before or after induction with Doxycycline for 48 h by anti-epitope Western blot. The * represents a cross-reacting band detected by the anti-HA antibody used as loading control. (B) Inducible expression of XPG-V5 from pLenti CMV/TO Zeo in U2OS cells monitored by Western blot (left) and immunofluorescence (right). In the whole population, very few cells (<1%) showed leakiness in the uninduced control; we photographed a field with some leaky uninduced cells. (C) LEFT: Western blot of Streptavidin pulldowns from HeLa S3 TREx-NTAP-p150 nuclear extracts (NE) without (lanes 1–4) or with (lanes 5–8) doxycycline induction (2 µg/ml) probed with anti-CBP (NTAP blot) or anti-p150. The distribution of NTAP-p150 during the nuclear extraction is the same as the endogenous p150, mainly absent from the cytosolic (S100) extract and present in the nuclear extract. The arrow represents NTAP-p150, where as the asterisk represents cross-reactivity of the antibody since it is in the uninduced samples and is not pelleted by the streptavidin beads (lane 8). RIGHT: Silver stain profile of purified NTAP-p150 (lane E) on a 5–20% gradient SDS-PAGE gel showing the p150, p60 and p48 subunits of CAF-1 migrating at their corresponding molecular weight (MW). BOTTOM: Immunofluorescence of NTAP-p150 shows that it is localized to replication foci as shown by its colocalization with PCNA.
Figure 8. Functional studies using the viral vectors.
(A) Inducible expression of GFP-TIN2-15C results in induction of DNA damage. GFP or GFP-iTIN2-15C were cloned into the pLenti CMV/TO Puro vector and transduced in HCA2 T-REx cells. Following a 96 h exposure to 0.1 ug/ml tetracycline, protein induction was monitored by GFP fluorescence and the presence of DNA damage was monitored by staining for 53BP1 foci. (B) Inducible expression or depletion of Asf1a in cells arrested in mitosis using the FKBP-DD system. Three experimental schemes are depicted with the corresponding lanes indicated by brackets. Lanes 1–6: Cells were treated with DMSO as a negative control. Lanes 7–12: Nocodazole treated cells. Lane 13: untreated cells. (C) Functional domains of the ARID4B protein. ARID4B is an 1312 amino acid protein with a Tudor domain (T) at its N-terminus, two repression domains ARID/R1 and R2 , and a chromo domain (shown in red). Two splice isoforms have been described and the chromo domain is spliced out in the shorter isoform. (D) The chromo domain of ARID4B is not required for G1-induced growth arrest. U2OS cells were transduced with either an untagged, V5-tagged or V5-tagged minus chromo version of ARID4B recombined in the pQCXP Destination vector. An empty vector was used as a negative control. G1 arrest was monitored by incubating the cells with nocodazole as described in Materials and Methods. The ARID4B Δchromo can still induce a G1 arrest indicating that the chromo domain is not required. On the right is shown the Western blot from the same experiment to monitor expression of each construct. (E) Localization of ARID4B and ARID4B Δchromo to the cell nucleus. The V5-tagged ARID4B isoforms were detected by indirect immunofluorescence using the V5 antibody and an anti-mouse Cy5 secondary antibody. Both isoforms are detected in the nucleus of U2OS cells.
Similar articles
- Construction of simple and efficient DNA vector-based short hairpin RNA expression systems for specific gene silencing in mammalian cells.
Cheng TL, Chang WT. Cheng TL, et al. Methods Mol Biol. 2007;408:223-41. doi: 10.1007/978-1-59745-547-3_13. Methods Mol Biol. 2007. PMID: 18314586 - High-throughput gateway bicistronic retroviral vectors for stable expression in mammalian cells: exploring the biologic effects of STAT5 overexpression.
Royer Y, Menu C, Liu X, Constantinescu SN. Royer Y, et al. DNA Cell Biol. 2004 Jun;23(6):355-65. doi: 10.1089/104454904323145245. DNA Cell Biol. 2004. PMID: 15231069 - A retroviral vector for siRNA expression in mammalian cells.
Størvold GL, Gjernes E, Askautrud HA, Børresen-Dale AL, Perou CM, Frengen E. Størvold GL, et al. Mol Biotechnol. 2007 Mar;35(3):275-82. doi: 10.1007/BF02686013. Mol Biotechnol. 2007. PMID: 17652791 - New experimental approaches in retrovirus-mediated expression screening.
Kitamura T. Kitamura T. Int J Hematol. 1998 Jun;67(4):351-9. doi: 10.1016/s0925-5710(98)00025-5. Int J Hematol. 1998. PMID: 9695408 Review. - miRNA cassettes in viral vectors: problems and solutions.
Liu YP, Berkhout B. Liu YP, et al. Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):732-45. doi: 10.1016/j.bbagrm.2011.05.014. Epub 2011 Jun 7. Biochim Biophys Acta. 2011. PMID: 21679781 Review.
Cited by
- Acute GARP depletion disrupts vesicle transport, leading to severe defects in sorting, secretion, and O-glycosylation.
Khakurel A, Pokrovskaya I, Lupashin VV. Khakurel A, et al. bioRxiv [Preprint]. 2024 Oct 14:2024.10.07.617053. doi: 10.1101/2024.10.07.617053. bioRxiv. 2024. PMID: 39416116 Free PMC article. Preprint. - Enhancement of Triple-Negative Breast Cancer-Specific Induction of Cell Death by Silver Nanoparticles by Combined Treatment with Proteotoxic Stress Response Inhibitors.
Snyder CM, Mateo B, Patel K, Fahrenholtz CD, Rohde MM, Carpenter R, Singh RN. Snyder CM, et al. Nanomaterials (Basel). 2024 Sep 27;14(19):1564. doi: 10.3390/nano14191564. Nanomaterials (Basel). 2024. PMID: 39404291 Free PMC article. - Indomethacin inhibits human seasonal coronaviruses at late stages of viral replication in lung cells: Impact on virus-induced COX-2 expression.
Tramontozzi C, Riccio A, Pauciullo S, La Frazia S, Rossi A, Santoro MG. Tramontozzi C, et al. J Virus Erad. 2024 Sep 1;10(3):100387. doi: 10.1016/j.jve.2024.100387. eCollection 2024 Sep. J Virus Erad. 2024. PMID: 39399815 Free PMC article. - Determination of Site-Specific Phosphorylation Occupancy Using Targeted Mass Spectrometry Reveals the Regulation of Human Apical Bile Acid Transporter, ASBT.
Nguyen TT, Kane MA, Swaan PW. Nguyen TT, et al. ACS Omega. 2024 Aug 30;9(37):38477-38489. doi: 10.1021/acsomega.4c02999. eCollection 2024 Sep 17. ACS Omega. 2024. PMID: 39310206 Free PMC article. - Purified CDT toxins and a clean deletion within the CDT locus provide novel insights into the contribution of binary toxin in cellular inflammation and Clostridioides difficile infection.
Nabukhotna K, Kordus SL, Shupe JA, Cano Rodríguez R, Smith A, Bohannon JK, Washington MK, Lacy DB. Nabukhotna K, et al. PLoS Pathog. 2024 Sep 19;20(9):e1012568. doi: 10.1371/journal.ppat.1012568. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39298531 Free PMC article.
References
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG017242/AG/NIA NIH HHS/United States
- F32 GM076863/GM/NIGMS NIH HHS/United States
- U54 CA112970/CA/NCI NIH HHS/United States
- P01 AG017242/AG/NIA NIH HHS/United States
- R01 CA063503/CA/NCI NIH HHS/United States
- 5 F32 GM076863-03/GM/NIGMS NIH HHS/United States
- R01 GM055712-11/GM/NIGMS NIH HHS/United States
- 1F32CA108393/CA/NCI NIH HHS/United States
- R01 GM055712/GM/NIGMS NIH HHS/United States
- R01 GM557/GM/NIGMS NIH HHS/United States
- P01 CA092584/CA/NCI NIH HHS/United States
- F32 CA108393/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous