The Neandertal genome and ancient DNA authenticity - PubMed (original) (raw)
Review
The Neandertal genome and ancient DNA authenticity
Richard E Green et al. EMBO J. 2009.
Abstract
Recent advances in high-thoughput DNA sequencing have made genome-scale analyses of genomes of extinct organisms possible. With these new opportunities come new difficulties in assessing the authenticity of the DNA sequences retrieved. We discuss how these difficulties can be addressed, particularly with regard to analyses of the Neandertal genome. We argue that only direct assays of DNA sequence positions in which Neandertals differ from all contemporary humans can serve as a reliable means to estimate human contamination. Indirect measures, such as the extent of DNA fragmentation, nucleotide misincorporations, or comparison of derived allele frequencies in different fragment size classes, are unreliable. Fortunately, interim approaches based on mtDNA differences between Neandertals and current humans, detection of male contamination through Y chromosomal sequences, and repeated sequencing from the same fossil to detect autosomal contamination allow initial large-scale sequencing of Neandertal genomes. This will result in the discovery of fixed differences in the nuclear genome between Neandertals and current humans that can serve as future direct assays for contamination. For analyses of other fossil hominins, which may become possible in the future, we suggest a similar 'boot-strap' approach in which interim approaches are applied until sufficient data for more definitive direct assays are acquired.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
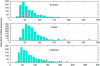
Figure 1
Estimates of human mtDNA contamination in Neandertal extracts. DNA extracts of Neandertal bones contain a large excess of microbial DNA (brown), at most a few percent of Neandertal DNA (blue) and generally variable amounts of contaminating DNA from current humans (red). Traditionally, contamination has been assayed through PCR directly from DNA extract from fossil bone (left lower panel). Accumulation of large numbers of reads from high-throughput sequencing allows a direct estimate of mtDNA contamination in the sequencing library (right lower panel). Once human/Neandertal diagnostic nuclear genome positions are learned, this strategy can be extended to nuclear DNA sequences.
Figure 2
Lengths of Neandertal and human mtDNA fragments. Distributions of mtDNA fragments carrying Neandertal diagnostic positions are shown in blue for three Neandertal fossils. Each red dot represents a single contaminating human mtDNA fragment of the indicated length (data from Briggs et al, 2009).
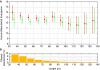
Figure 3
Neandertal/human divergence estimated from sequences of increasing length and score filtering. (A) Sequences in each length bin were used to calculate human/Neandertal divergence, given as the percentage of the human lineage back to the human/chimpanzee common ancestor in which the Neandertal sequences diverged. Sequences were filtered for uniqueness in the human and chimpanzee genomes by comparing the best alignment score to the second best score. In red are sequences whose best alignments are at least 1-bit better than the second best, in green with a difference of 5 bits or more. Bars show the 95% confidence interval from 1000 bootstrap replicates of the sequences in each bin. (B) Percentage of the sequences in each bin removed when increasing the alignment score filter from 1 to 5 bits. Shorter sequences are more likely to be removed by stricter filtering as they carry less information to place them uniquely in the human and chimpanzee genomes.
Figure 4
Fraction of human polymorphic positions carrying derived alleles in Neandertal and human DNA sequences. (A) Neandertal sequences of increasing length that overlap human polymorphic positions were assessed for having the derived or ancestral (chimpanzee-like) allele. Blue points are for Neandertal data, red points for the corresponding sequences in the human reference genome (hg18). (B) Sequences of length 60–78 nucleotides were split in half and re-analysed (‘30–39'). Derived alleles are preferentially lost when fragments size is reduced.
Similar articles
- Targeted retrieval and analysis of five Neandertal mtDNA genomes.
Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Schmitz R, Doronichev VB, Golovanova LV, de la Rasilla M, Fortea J, Rosas A, Pääbo S. Briggs AW, et al. Science. 2009 Jul 17;325(5938):318-21. doi: 10.1126/science.1174462. Science. 2009. PMID: 19608918 - Neandertal DNA sequences and the origin of modern humans.
Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Krings M, et al. Cell. 1997 Jul 11;90(1):19-30. doi: 10.1016/s0092-8674(00)80310-4. Cell. 1997. PMID: 9230299 - A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing.
Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, Prüfer K, Siebauer M, Burbano HA, Ronan M, Rothberg JM, Egholm M, Rudan P, Brajković D, Kućan Z, Gusić I, Wikström M, Laakkonen L, Kelso J, Slatkin M, Pääbo S. Green RE, et al. Cell. 2008 Aug 8;134(3):416-26. doi: 10.1016/j.cell.2008.06.021. Cell. 2008. PMID: 18692465 Free PMC article. - Ancient DNA: would the real Neandertal please stand up?
Cooper A, Drummond AJ, Willerslev E. Cooper A, et al. Curr Biol. 2004 Jun 8;14(11):R431-3. doi: 10.1016/j.cub.2004.05.037. Curr Biol. 2004. PMID: 15182692 Review. - [Progresses on Neandertal genomics].
Bi CL, Guo GY, Zhang X, Tian YH, Shen YZ. Bi CL, et al. Yi Chuan. 2012 Jun;34(6):659-65. doi: 10.3724/sp.j.1005.2012.00659. Yi Chuan. 2012. PMID: 22698735 Review. Chinese.
Cited by
- SAFARI: Pangenome Alignment of Ancient DNA Using Purine/Pyrimidine Encodings.
Rubin J, van Waaij J, Kraft L, Sirén J, Sackett PW, Renaud G. Rubin J, et al. bioRxiv [Preprint]. 2024 Oct 8:2024.08.12.607489. doi: 10.1101/2024.08.12.607489. bioRxiv. 2024. PMID: 39415996 Free PMC article. Preprint. - Evaluation of the usefulness of insertion-null markers in critical skeletal remains.
Haarkötter C, Saiz M, Gálvez X, Vinueza-Espinosa DC, Medina-Lozano MI, Álvarez JC, Lorente JA. Haarkötter C, et al. Int J Legal Med. 2024 Jul;138(4):1287-1293. doi: 10.1007/s00414-024-03205-3. Epub 2024 Mar 20. Int J Legal Med. 2024. PMID: 38509248 Free PMC article. - Developing the Protocol Infrastructure for DNA Sequencing Natural History Collections.
Ferrari G, Esselens L, Hart ML, Janssens S, Kidner C, Mascarello M, Peñalba JV, Pezzini F, von Rintelen T, Sonet G, Vangestel C, Virgilio M, Hollingsworth PM. Ferrari G, et al. Biodivers Data J. 2023 Oct 27;11:e102317. doi: 10.3897/BDJ.11.e102317. eCollection 2023. Biodivers Data J. 2023. PMID: 38327316 Free PMC article. - Review: Computational analysis of human skeletal remains in ancient DNA and forensic genetics.
Childebayeva A, Zavala EI. Childebayeva A, et al. iScience. 2023 Oct 4;26(11):108066. doi: 10.1016/j.isci.2023.108066. eCollection 2023 Nov 17. iScience. 2023. PMID: 37927550 Free PMC article. Review. - Density separation of petrous bone powders for optimized ancient DNA yields.
Fernandes DM, Sirak KA, Cheronet O, Novak M, Brück F, Zelger E, Llanos-Lizcano A, Wagner A, Zettl A, Mandl K, Duffet Carlson KS, Oberreiter V, Özdoğan KT, Sawyer S, La Pastina F, Borgia E, Coppa A, Dobeš M, Velemínský P, Reich D, Bell LS, Pinhasi R. Fernandes DM, et al. Genome Res. 2023 Apr;33(4):622-631. doi: 10.1101/gr.277714.123. Epub 2023 Apr 18. Genome Res. 2023. PMID: 37072186 Free PMC article.
References
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ et al. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 - PMC - PubMed
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Pavao R, Brajkovic D, Kucan Z, Gusic I, Schmitz RW, Doronichev VB, Golovanova LV, de la Resilla M, Fortea J, Rosas A, Pääbo S (2009) Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325: 318–321 - PubMed
- Brown P, Sutikna T, Morwood MJ, Soejono RP, Jatmiko, Saptomo EW, Due RA (2004) A new small-bodied hominin from the Late Pleistocene of Flores, Indonesia. Nature 431: 1055–1061 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources