Chromatin architecture and the generation of antigen receptor diversity - PubMed (original) (raw)
Review
Chromatin architecture and the generation of antigen receptor diversity
Suchit Jhunjhunwala et al. Cell. 2009.
Abstract
The adaptive immune system generates a specific response to a vast spectrum of antigens. This remarkable property is achieved by lymphocytes that each express single and unique antigen receptors. During lymphocyte development, antigen receptor coding elements are assembled from widely dispersed gene segments. The assembly of antigen receptors is controlled at multiple levels, including epigenetic marking, nuclear location, and chromatin topology. Here, we review recently uncovered mechanisms that underpin long-range genomic interactions and the generation of antigen receptor diversity.
Figures
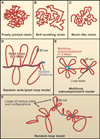
Figure 1. Flexible Polymer Chain Models in Free and Confined Environments
(A) Freely jointed chain. A freely jointed chain consists of a series of rigid segments connected by flexible hinges. (B) Self avoiding random walk. In a self-avoiding chain, a segment cannot intersect any other segment. (C) Worm-like chain. In contrast to the freely-jointed chain, which is flexible within the hinges that separate the segments, the worm-like polymer chain is continuously flexible. (D) Random walk/giant loop model. Giant loops of 3–5 Mbp are tethered to a backbone. The DNA within the loops and the backbone itself follow a random walk. (E) Multi-loop subcompartment model. Chromatin is organized into 1–2 Mbp subcompartments. Each subcompartment consists of a bundle of loops that are attached to a common loop base. Linkers connect the chromatin subcompartments. Both loops and linkers undergo random walk behavior. (F) Random loop model. Dynamic loops of large and small sizes are formed at random intervals on the chromosome. Both individual loops and bundles of loops are shown.
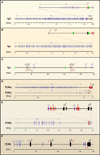
Figure 2. Genomic Organization of Antigen Receptor Loci
Genetic structures of antigen receptor loci are shown. Variable (V) gene segments are shown in blue; Diversity (D) gene segments, if present in purple; Joining (J) gene segments in red; Constant © regions a in black; Enhancers in green. Note that the TCRδ locus is interspersed within the TCRα locus. Genomic distances (Mbp or Kbp) are indicated for each of the loci.
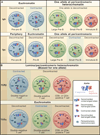
Figure 3. Location and Conformation of Antigen Receptor Loci in Developing Lymphocytes
Nuclear locations of antigen receptor loci during developmental progression are indicated. (A) Indicated are the nuclear positions of the Igh and Igκ alleles at various stages of early B cell development. Pre-pro-B and pro-B cell stage are shown in blue. Large pre-B and small pre-B are indicated in green. Immature-B cells are shown in red. Blue cluster of loops indicate V regions. Red clusters of loops represent D/J/C coding elements. Dark dots represent the pericentromeric heterochromatin. Relative degree of Igh and Igκ locus contraction and de-contraction is depicted. (B) Indicated are the nuclear positions of the TCRβ and TCRα alleles during thymopoieisis. The double negative cell stage is shown in blue. Double positive cells are in green; single positive cells in red. Blue clusters of loops indicate V regions. Red clusters of loops represent D/J/C coding elements. Dark dots represent the pericentromeric heterochromatin. Relative degree of TCRα and TCRβ contraction and de-contraction are depicted.
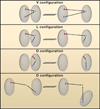
Figure 4. Genomic Elements and Chromatin Territories
Positioning of genomic markers in distinct compartments using triple point spatial distance measurements in individual cells. Two genomic markers are located in one compartment if they move coordinately to or away from an anchor. Nuclear subcompartments are depicted as grey discs. Genomic markers within different subcompartments are shown in red, green, or blue. Spatial distances from the anchor (green) are shown as red or blue, respectively. Various configurations are indicated. V-configuration; Red and blue markers are in the same subcompartment whereas anchor (green) is positioned in a separate subcompartment. The red and blue markers move coordinately away from the anchor. L-configuration; One of the markers (blue) is in the same subcompartment as the anchor (green). The distance separating the blue marker from the anchor is independent of the distance separating the red marker from the anchor. O-configuration; All three genomic markers are in localized in one compartment or alternatively in three distinct compartments. The markers move independently from each other.
Similar articles
- Chromatin topology and the regulation of antigen receptor assembly.
Bossen C, Mansson R, Murre C. Bossen C, et al. Annu Rev Immunol. 2012;30:337-56. doi: 10.1146/annurev-immunol-020711-075003. Epub 2012 Jan 3. Annu Rev Immunol. 2012. PMID: 22224771 Review. - Epigenetics of antigen-receptor gene assembly.
Murre C. Murre C. Curr Opin Genet Dev. 2007 Oct;17(5):415-21. doi: 10.1016/j.gde.2007.08.006. Epub 2007 Oct 24. Curr Opin Genet Dev. 2007. PMID: 17920858 Free PMC article. Review. - Mechanisms of antigen receptor evolution.
Eason DD, Cannon JP, Haire RN, Rast JP, Ostrov DA, Litman GW. Eason DD, et al. Semin Immunol. 2004 Aug;16(4):215-26. doi: 10.1016/j.smim.2004.08.001. Semin Immunol. 2004. PMID: 15522620 Review. - Receptor editing in lymphocyte development and central tolerance.
Nemazee D. Nemazee D. Nat Rev Immunol. 2006 Oct;6(10):728-40. doi: 10.1038/nri1939. Nat Rev Immunol. 2006. PMID: 16998507 Review. - Variable Lymphocyte Receptors: A Current Overview.
Kasahara M. Kasahara M. Results Probl Cell Differ. 2015;57:175-92. doi: 10.1007/978-3-319-20819-0_8. Results Probl Cell Differ. 2015. PMID: 26537382 Review.
Cited by
- Subnuclear cyclin D3 compartments and the coordinated regulation of proliferation and immunoglobulin variable gene repression.
Powers SE, Mandal M, Matsuda S, Miletic AV, Cato MH, Tanaka A, Rickert RC, Koyasu S, Clark MR. Powers SE, et al. J Exp Med. 2012 Nov 19;209(12):2199-213. doi: 10.1084/jem.20120800. Epub 2012 Oct 29. J Exp Med. 2012. PMID: 23109711 Free PMC article. - Highly diverse TCRα chain repertoire of pre-immune CD8⁺ T cells reveals new insights in gene recombination.
Genolet R, Stevenson BJ, Farinelli L, Osterås M, Luescher IF. Genolet R, et al. EMBO J. 2012 Apr 4;31(7):1666-78. doi: 10.1038/emboj.2012.48. Epub 2012 Feb 28. EMBO J. 2012. PMID: 22373576 Free PMC article. - Deciphering the Complexity of 3D Chromatin Organization Driving Lymphopoiesis and Lymphoid Malignancies.
Scourzic L, Salataj E, Apostolou E. Scourzic L, et al. Front Immunol. 2021 May 14;12:669881. doi: 10.3389/fimmu.2021.669881. eCollection 2021. Front Immunol. 2021. PMID: 34054841 Free PMC article. Review. - Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark.
Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Criscitiello MF, et al. J Immunol. 2010 Jun 15;184(12):6950-60. doi: 10.4049/jimmunol.0902774. Epub 2010 May 19. J Immunol. 2010. PMID: 20488795 Free PMC article. - Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination.
Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Ji Y, et al. J Exp Med. 2010 Dec 20;207(13):2809-16. doi: 10.1084/jem.20101136. Epub 2010 Nov 29. J Exp Med. 2010. PMID: 21115692 Free PMC article.
References
- Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat. Immunol. 2006;7:1109–1115. - PubMed
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor beta gene rearrangements and allelic exclusion. Immunity. 2007;27:871–884. - PubMed
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources