The autophagy machinery is required to initiate hepatitis C virus replication - PubMed (original) (raw)
The autophagy machinery is required to initiate hepatitis C virus replication
Marlène Dreux et al. Proc Natl Acad Sci U S A. 2009.
Abstract
In addition to its cellular homeostasis function, autophagy is emerging as a central component of antimicrobial host defense against diverse infections. To counteract this mechanism, many pathogens have evolved to evade, subvert, or exploit autophagy. Here, we report that autophagy proteins (i.e., Beclin-1, Atg4B, Atg5, and Atg12) are proviral factors required for translation of incoming hepatitis C virus (HCV) RNA and, thereby, for initiation of HCV replication, but they are not required once infection is established. These results illustrate a previously unappreciated role for autophagy in the establishment of a viral infection and they suggest that different host factors regulate the translation of incoming viral genome and translation of progeny HCV RNA once replication is established.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
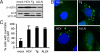
Enhanced autophagic vesicle content in HCV-infected cells. (A) Analysis by immnunoblotting of endogenous LC3 lipidation in cell extracts of Huh-7 cells infected for 24 h with HCV or treated with thapsigargin (Tg) (500 nM) or with ALLN (10 μM) for 16 h, or untreated (mock), as indicated. β-actin expression was examined as a protein loading control. The gels are representative of three independent experiments. (B) Representative confocal images. LC3-GFP transfected Huh-7 cells were grown in normal medium (mock), infected for 24 h with HCV, or treated with ALLN or with Tg, as described in A. The nuclei were stained by Hoechst solution (blue). (C) Quantification of the frequency of Huh-7 cells displaying a punctate distribution of LC3-GFP. Results represent the means of two independent experiments.
Fig. 2.
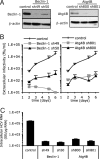
Autophagy machinery is required for HCV spread. Huh-7 cells were transduced with lentiviral vectors expressing shRNA against Beclin-1 (shRNA denoted as sh49 and sh50) or Atg4B (sh800 and sh801). The selected shRNA sequence and multiplicity of infection (MOI) used to transduce Huh-7 cells did not compromise the cell viability. At 8 days posttransduction, transduced and control cells were harvested and seeded in equal numbers. Twenty and seventy-two hours later MTT was added and cellular proliferation was monitored. The results for the Beclin-1-(sh49) transduced cells were 99.3 ± 27.3% of the nontransduced cells, set to 100. (A) The relative abundance of Beclin-1 and Atg4B proteins in the Huh-7 cells was analyzed by immunoblotting. β-actin expression was used as a protein loading control. (B) Beclin-1- and Atg4B-deficient and nontransduced (control) Huh-7 cells were infected with HCV at an MOI of 0.01. The infectivity of their virus-containing cell supernatants was determined at different times postinoculation, as indicated. Results display average infectious titers, expressed as focus-forming units (ffu)/ml (mean ± SD; n = 2). (C) The intracellular HCV RNA levels were determined by RT-qPCR 6 days postinoculation (MOI of 0.01) in Beclin-1- and Atg4B-deficient and control cells (mean ± SD; n = 2). GE, genome equivalent.
Fig. 3.
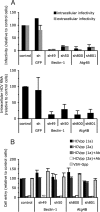
The autophagy machinery regulates a postentry step, upstream or at the level of HCV replication. Huh-7 cells were transduced or not (control) with lentiviral vectors expressing shRNA against Beclin-1, Atg4B, and GFP. (A) Extracellular and intracellular infectious virus and intracellular RNA production during 24 h in a single step infection at MOI of 10 (mean ± SD; n = 3). (Upper) Accumulation of extracellular and intracellular HCV infectious particles was determined 24 h after inoculation, and expressed as a percentage of that in control cells. (Lower) Intracellular HCV RNA content was determined at 24 h post inoculation and expressed as a percentage of that in control cells. (B) Cell entry of HCVpp harboring E1E2 glycoproteins derived from HCV strains H77 (1a) or JHF-1 (2a) and control particles harboring the VSV-G glycoprotein (VSV-Gpp) expressed as a percentage of that in control cells (mean ± SD; n = 4). To assess the specificity of E1E2 glycoprotein-mediated cell entry, anti-E2 antibody (Ab) was added at 20 μg/mL. ND, not determined.
Fig. 4.
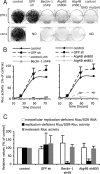
Autophagy machinery regulates translation and/or delivery of incoming viral RNA to the translation apparatus. Huh-7 cells were transduced with lentivirus expressing shRNA against Beclin-1, Atg4B, and GFP. (A) Analysis of JFH1 and Con1 neo/SGR replication. Neomycin selected cells were fixed and stained with Crystal Violet. A replication defective GND mutant (GDD-to-GND mutation in the NS5B protein) neo/SGR was used as a positive control for the neomycin selection. Results are representative of two independent experiments. (B) Analysis of Rluc/SGR translation/replication by monitoring Rluc activity at different times posttransfection. To assess the specificity of Rluc activity, control cells were treated with 5 μM of 2′-C-methyladenosine, a HCV polymerase inhibitor (denoted inh). For each independent experiment, Rluc activity was normalized to cell density and expressed as a percentage of that determined in control cells at 72 h posttransfection (mean ± SD; n = 4). (C) Translation of replication-deficient Rluc/SGR RNAs. Intracellular RNA levels and Rluc activity of replication deficient Rluc/SGR were determined at 6 h posttransfection. Rluc activities are statistically reduced in autophagy protein deficient cells compared to shRNA GFP expressing cells (P values <0.05 from paired Student's t test). In parallel transfections, Rluc activity expressed from pRL-TK plasmid (irrelevant-Rluc activity) was determined. For each independent experiment, Rluc activity was normalized to cell density and expressed as a percentage of that determined in control cells (mean ± SD; n = 3).
Fig. 5.
Autophagy machinery does not regulate established HCV replication. (A) Replicon bearing-Huh-7 cells were transduced with lentiviral vectors expressing shRNAs against Beclin-1, Atg4B, and GFP. Protein and RNA levels were determined 10 days posttransduction. (Upper Left) NS5A protein levels in replicon cells as analyzed by immunoblotting. (Upper Right) Intracellular HCV RNA levels in H77 (1a), con-1 (1b), JFH-1 (2a) SGR, or JFH-1 full-length (FLR) replicon-bearing Huh-7 cells monitored by RT-qPCR (mean ± SD; n = 2). GE, genome equivalent. (Lower) Efficiency of down-regulation of Beclin-1, Atg4B, as analyzed by immunoblotting. Beclin-1 and Atg4B content were analyzed in established JFH-1 SGR replicon cells (denoted Replicon cells) and were compared to shRNA-treated cells before HCV infection (denoted DR before infection). β-actin expression was used as a protein loading control. (B) Huh-7 cells that were virtually all infected by HCV were transduced with lentiviral vectors expressing shRNA against Beclin-1, Atg4B. (Left) Relative levels of Beclin-1 and Atg4B in the infected cells, as analyzed by immunoblotting. β-actin expression was used as a protein loading control. (Right) Extracellular and intracellular infectivity and intracellular HCV RNA levels were determined 8 days posttransduction and are expressed as a percentage of those in control cells (mean ± SD; n = 2).
Similar articles
- Regulation of Autophagy by Hepatitis C Virus for Its Replication.
Wang L, Ou JJ. Wang L, et al. DNA Cell Biol. 2018 Apr;37(4):287-290. doi: 10.1089/dna.2017.4115. Epub 2018 Jan 19. DNA Cell Biol. 2018. PMID: 29350547 Free PMC article. Review. - Perturbation of autophagic pathway by hepatitis C virus.
Sir D, Liang C, Chen WL, Jung JU, Ou JH. Sir D, et al. Autophagy. 2008 Aug;4(6):830-1. doi: 10.4161/auto.6566. Epub 2008 Jul 8. Autophagy. 2008. PMID: 18635950 Free PMC article. - Autophagy proteins promote hepatitis C virus replication.
Dreux M, Chisari FV. Dreux M, et al. Autophagy. 2009 Nov;5(8):1224-5. doi: 10.4161/auto.5.8.10219. Epub 2009 Nov 1. Autophagy. 2009. PMID: 19844160 - Impact of the autophagy machinery on hepatitis C virus infection.
Dreux M, Chisari FV. Dreux M, et al. Viruses. 2011 Aug;3(8):1342-57. doi: 10.3390/v3081342. Epub 2011 Aug 4. Viruses. 2011. PMID: 21994783 Free PMC article. Review. - Stem-loop structures II-IV of the 5' untranslated sequences are required for the expression of the full-length hepatitis C virus genome.
Qi ZT, Kalkeri G, Hanible J, Prabhu R, Bastian F, Garry RF, Dash S. Qi ZT, et al. Arch Virol. 2003 Mar;148(3):449-67. doi: 10.1007/s00705-002-0933-0. Arch Virol. 2003. PMID: 12607098
Cited by
- Regulation of hepatic innate immunity by hepatitis C virus.
Horner SM, Gale M Jr. Horner SM, et al. Nat Med. 2013 Jul;19(7):879-88. doi: 10.1038/nm.3253. Nat Med. 2013. PMID: 23836238 Free PMC article. Review. - Guanylate-binding protein 1 acts as a pro-viral factor for the life cycle of hepatitis C virus.
Bender D, Koulouri A, Wen X, Glitscher M, Schollmeier A, Fernandes da Costa L, Murra RO, Carra GP, Haberger V, Praefcke GJK, Hildt E. Bender D, et al. PLoS Pathog. 2024 Feb 5;20(2):e1011976. doi: 10.1371/journal.ppat.1011976. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38315728 Free PMC article. - Lyn kinase regulates egress of flaviviruses in autophagosome-derived organelles.
Li MY, Naik TS, Siu LYL, Acuto O, Spooner E, Wang P, Yang X, Lin Y, Bruzzone R, Ashour J, Evans MJ, Sanyal S. Li MY, et al. Nat Commun. 2020 Oct 15;11(1):5189. doi: 10.1038/s41467-020-19028-w. Nat Commun. 2020. PMID: 33060596 Free PMC article. - Targeting autophagy for the treatment of liver diseases.
Ni HM, Williams JA, Yang H, Shi YH, Fan J, Ding WX. Ni HM, et al. Pharmacol Res. 2012 Dec;66(6):463-74. doi: 10.1016/j.phrs.2012.07.003. Epub 2012 Jul 31. Pharmacol Res. 2012. PMID: 22871337 Free PMC article. Review. - Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells.
Zeng X, Carlin CR. Zeng X, et al. J Virol. 2013 Feb;87(4):2307-19. doi: 10.1128/JVI.02014-12. Epub 2012 Dec 12. J Virol. 2013. PMID: 23236070 Free PMC article.
References
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. - PubMed
- Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases