Application of massive parallel sequencing to whole genome SNP discovery in the porcine genome - PubMed (original) (raw)
Application of massive parallel sequencing to whole genome SNP discovery in the porcine genome
Andreia J Amaral et al. BMC Genomics. 2009.
Abstract
Background: Although the Illumina 1 G Genome Analyzer generates billions of base pairs of sequence data, challenges arise in sequence selection due to the varying sequence quality. Therefore, in the framework of the International Porcine SNP Chip Consortium, this pilot study aimed to evaluate the impact of the quality level of the sequenced bases on mapping quality and identification of true SNPs on a large scale.
Results: DNA pooled from five animals from a commercial boar line was digested with DraI; 150-250-bp fragments were isolated and end-sequenced using the Illumina 1 G Genome Analyzer, yielding 70,348,064 sequences 36-bp long. Rules were developed to select sequences, which were then aligned to unique positions in a reference genome. Sequences were selected based on quality, and three thresholds of sequence quality (SQ) were compared. The highest threshold of SQ allowed identification of a larger number of SNPs (17,489), distributed widely across the pig genome. In total, 3,142 SNPs were validated with a success rate of 96%. The correlation between estimated minor allele frequency (MAF) and genotyped MAF was moderate, and SNPs were highly polymorphic in other pig breeds. Lowering the SQ threshold and maintaining the same criteria for SNP identification resulted in the discovery of fewer SNPs (16,768), of which 259 were not identified using higher SQ levels. Validation of SNPs found exclusively in the lower SQ threshold had a success rate of 94% and a low correlation between estimated MAF and genotyped MAF. Base change analysis suggested that the rate of transitions in the pig genome is likely to be similar to that observed in humans. Chromosome X showed reduced nucleotide diversity relative to autosomes, as observed for other species.
Conclusion: Large numbers of SNPs can be identified reliably by creating strict rules for sequence selection, which simultaneously decreases sequence ambiguity. Selection of sequences using a higher SQ threshold leads to more reliable identification of SNPs. Lower SQ thresholds can be used to guarantee sufficient sequence coverage, resulting in high success rate but less reliable MAF estimation. Nucleotide diversity varies between porcine chromosomes, with the X chromosome showing less variation as observed in other species.
Figures
Figure 1
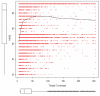
Maximum mapping quality (MMQ) (mapping quality of the best mapped sequence of a cluster) on an SNP position versus target coverage. Box plots show the data distribution for each parameter. Red dots show MMQ values for the best mapped sequence on an SNP position versus target coverage. The black solid line shows the smooth-fit line.
Figure 2
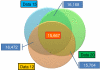
Venn diagram showing the number of identical SNPs between the analyzed data sets with different levels of sequence quality.
Figure 3
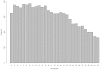
Number of identified SNPs per position in a short read for Data 20.
Figure 4
SNP map of each chromosome based on Data 20. The colored vertical lines represent the location of each SNP.
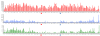
Figure 5
Sequence coverage, nucleotide diversity, and SNP occurrence along chromosome 1. Each bar represents a window of 1 Mb. Red bars show the length of the aligned consensus sequence, blue bars show the estimated level of nucleotide diversity, and green bars show the number of SNPs found in each window. The red triangle designates the position of the centromere. The blue triangle designates a position where nucleotide diversity is high where coverage is low.
Similar articles
- The development and characterization of a 60K SNP chip for chicken.
Groenen MA, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RP, Vereijken A, Okimoto R, Muir WM, Cheng HH. Groenen MA, et al. BMC Genomics. 2011 May 31;12(1):274. doi: 10.1186/1471-2164-12-274. BMC Genomics. 2011. PMID: 21627800 Free PMC article. - SNP discovery in swine by reduced representation and high throughput pyrosequencing.
Wiedmann RT, Smith TP, Nonneman DJ. Wiedmann RT, et al. BMC Genet. 2008 Dec 4;9:81. doi: 10.1186/1471-2156-9-81. BMC Genet. 2008. PMID: 19055830 Free PMC article. - Large scale single nucleotide polymorphism discovery in unsequenced genomes using second generation high throughput sequencing technology: applied to turkey.
Kerstens HH, Crooijmans RP, Veenendaal A, Dibbits BW, Chin-A-Woeng TF, den Dunnen JT, Groenen MA. Kerstens HH, et al. BMC Genomics. 2009 Oct 16;10:479. doi: 10.1186/1471-2164-10-479. BMC Genomics. 2009. PMID: 19835600 Free PMC article. - Analyses of porcine public SNPs in coding-gene regions by re-sequencing and phenotypic association studies.
Li X, Kim SW, Do KT, Ha YK, Lee YM, Yoon SH, Kim HB, Kim JJ, Choi BH, Kim KS. Li X, et al. Mol Biol Rep. 2011 Aug;38(6):3805-20. doi: 10.1007/s11033-010-0496-1. Epub 2010 Nov 24. Mol Biol Rep. 2011. PMID: 21107721 - Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa).
Chen K, Baxter T, Muir WM, Groenen MA, Schook LB. Chen K, et al. Int J Biol Sci. 2007 Feb 10;3(3):153-65. doi: 10.7150/ijbs.3.153. Int J Biol Sci. 2007. PMID: 17384734 Free PMC article. Review.
Cited by
- Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary.
Molnár J, Nagy T, Stéger V, Tóth G, Marincs F, Barta E. Molnár J, et al. BMC Genomics. 2014 Sep 5;15(1):761. doi: 10.1186/1471-2164-15-761. BMC Genomics. 2014. PMID: 25193519 Free PMC article. - Recombination of the porcine X chromosome: a high density linkage map.
Fernández AI, Muñoz M, Alves E, Folch JM, Noguera JL, Enciso MP, Rodríguez Mdel C, Silió L. Fernández AI, et al. BMC Genet. 2014 Dec 20;15:148. doi: 10.1186/s12863-014-0148-x. BMC Genet. 2014. PMID: 25526890 Free PMC article. - Genome wide SNP discovery, analysis and evaluation in mallard (Anas platyrhynchos).
Kraus RH, Kerstens HH, Van Hooft P, Crooijmans RP, Van Der Poel JJ, Elmberg J, Vignal A, Huang Y, Li N, Prins HH, Groenen MA. Kraus RH, et al. BMC Genomics. 2011 Mar 16;12:150. doi: 10.1186/1471-2164-12-150. BMC Genomics. 2011. PMID: 21410945 Free PMC article. - Mutation discovery for Mendelian traits in non-laboratory animals: a review of achievements up to 2012.
Nicholas FW, Hobbs M. Nicholas FW, et al. Anim Genet. 2014 Apr;45(2):157-70. doi: 10.1111/age.12103. Epub 2013 Dec 26. Anim Genet. 2014. PMID: 24372556 Free PMC article. Review. - Genome-Wide SNP Discovery and Analysis of Genetic Diversity in Farmed Sika Deer (Cervus nippon) in Northeast China Using Double-Digest Restriction Site-Associated DNA Sequencing.
Ba H, Jia B, Wang G, Yang Y, Kedem G, Li C. Ba H, et al. G3 (Bethesda). 2017 Sep 7;7(9):3169-3176. doi: 10.1534/g3.117.300082. G3 (Bethesda). 2017. PMID: 28751500 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases