Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element - PubMed (original) (raw)
Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element
James R R Whittle et al. J Biol Chem. 2009.
Abstract
The nuclear pore complex (NPC) constitutes one of the largest protein assemblies in the eukaryotic cell and forms the exclusive gateway to the nucleus. The stable, approximately 15-20-MDa scaffold ring of the NPC is built from two multiprotein complexes arranged around a central 8-fold axis. Here we present crystal structures of two large architectural units, yNup170(979-1502) and hNup107(658-925) x hNup133(517-1156), each a constituent of one of the two multiprotein complexes. Conservation of domain arrangement and of tertiary structure suggests that Nup157/170 and Nup133 derived from a common ancestor. Together with the previously established ancestral coatomer element (ACE1), these two elements constitute the major alpha-helical building blocks of the NPC scaffold and define its branched, lattice-like architecture, similar to vesicle coats like COPII. We hypothesize that the extant NPC evolved early during eukaryotic evolution from a rudimentary structure composed of several identical copies of a few ancestral elements, later diversified and specified by gene duplication.
Figures
FIGURE 1.
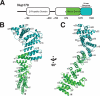
Nup170979–1502 forms a bipartite, irregular α-helical stack. A, the predicted domain structure of Nup170 is diagramed, with the β-propeller domain (residues 180–650) and α-helical domains (residues 760–1502) drawn as ovals. Indentation at residue 1270 reflects the division into two parts. The portion of the structure solved is colored as in B and C. B, cartoon representation of the 3.2-Å structure of the α-helical C-terminal domain of Nup170 with its two subdomains shown in green and aquamarine, respectively. Secondary structure elements and termini are labeled. Disordered loops connecting structural elements are drawn as dotted lines. A central, long helix, α14, spans and connects the two subdomains. The C-terminal subdomain, helices α14–26 was also expressed as a separate protein fragment and solved at 2.2-Å resolution. C, structure rotated by 90° compared with B.
FIGURE 2.
Nup133517–1156 in complex with Nup84(hNup107658–925). A, the domain structures of Nup107 and Nup133 are diagramed. Nup107 consists of a single α-helical domain homologous to other ACE1 proteins. Nup133 has a β-propeller domain (residues 76–478) solved previously and an α-helical domain as shown in B and C. The portions of each molecule solved here are colored. The indentation at residue 1008 of Nup133 reflects the hinge between helices α24 and α25. B, the α-helical domain of Nup133 (orange) in complex with Nup107658–925 (blue). Loops evident in the electron density, but not modeled for the purpose of refinement, are gray. Secondary structure elements and termini of Nup133 are labeled, beginning at the N terminus of the domain, helix α4. C, structure rotated by 90° compared with B. Nup107 helix α6′, a metazoan-specific structural element, is labeled.
FIGURE 3.
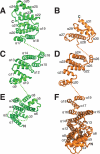
The topology of Nup170 and Nup133 α-helical domains are conserved. Nup170 in green and Nup133 in orange were compared by structural alignment of segmental subdomains. Nup170 N-terminal (E), central (C), and C-terminal (A) subdomains were structural aligned to the related segments (F, D, and B) in Nup133. Secondary structure elements are labeled. Unmodeled loops and connections between domains are shown as dotted lines. The N-terminal remainder of each molecule not solved here extends where marked N. The C terminus of each molecule is marked C.
FIGURE 4.
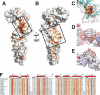
Surface conservation of Nup170 suggests two protein-protein interfaces. A, amino acid sequence conservation among Nup170 genes from maximally diverse eukaryotes was mapped on the protein, gradient-colored from white (not conserved) to orange (strongly conserved), orientated as in Fig. 1_B._ A conserved groove is boxed. B, structure rotated 180° compared with A, with conserved surface patch boxed. C, surface groove boxed in A shown magnified. Structure of C-terminal subdomain at 2.2-Å resolution is superposed and shown in aquamarine as a cartoon. Helix α14 as white cartoon, extending down and left, in the conformation seen in the full domain, as well as in aquamarine as the well ordered, extended peptide seen in the 2.2-Å structure of the isolated C-terminal subdomain. The key residues are labeled. D, surface patch boxed in B is magnified and shown as a cartoon with exposed residues labeled. Partially transparent surface representation is colored by calculated surface charge, in a gradient from negative (red) to neutral (white) to positive (blue). E, homologous section of Nup133 colored and labeled as in D with extent of interface to Nup107 delimited by a solid black line. F, sequence alignment of maximally diverse selection of eukaryotic Nup170 sequences, colored by conservation as in A–C. Helical segments are shown as red cylinders and labeled. The bar graph shows accessible surface area (Å2) for each residue. Yellow circles mark conserved, buried arginine 1232; two hydrophilic surface residues, glutamine 1234 and arginine 1238, that would be buried were this surface, shown in D, indeed a protein-protein interface, as in Nup133, shown in E; and residues serine 1305, phenylalanine 1308, and phenylalanine 1325, lining the groove shown in C.
FIGURE 5.
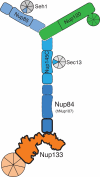
Schematic representation of the heptameric Nup84 subcomplex. The Nup84 subcomplex is composed of three ACE1 proteins (blues), Nup133 (orange), Nup120 (green), and β-propeller proteins Seh1 and Sec13 (gray). Nup85 and Nup145C each contribute in trans one blade of the Seh1 and Sec13 β-propellers. The portions of Nup133 and Nup107 solved here, as shown in Fig. 2, are outlined in bold. The N-terminal β-propeller of Nup133 and all portions of the Y outlined by solid lines have been solved at atomic resolution previously. The dashed lines denote portions of the Nup84 subcomplex for which no atomic resolution structures have yet been published.
FIGURE 6.
Representative nucleoporin α-helical stack domains. A, the overall topology of human Nup133 (residues 75–1156) is shown. The structure is gradient-colored from orange to white from the N terminus of the helical domain to the C terminus. The β-propeller domain is colored gray. B, the overall topology of yeast Nic96 (residues 200–835), an ACE1, is shown. The structure is gradient colored from red to white from N to C terminus. C, the overall topology of yeast Nup120 (residues 1–730 of 1037) is shown. The α-helical domain is gradient-colored from magenta to white from N to C terminus. Residue 1–381 are colored gray and form β-propeller blades β1-β6 and strand β7D. Note that strands A–C of blade β7 are contributed by the portion of the α-helical domain that links helix α4 to helix α5.
Similar articles
- Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Brohawn SG, et al. Science. 2008 Nov 28;322(5906):1369-73. doi: 10.1126/science.1165886. Epub 2008 Oct 30. Science. 2008. PMID: 18974315 Free PMC article. - Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review.
Cited by
- Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - A jumbo problem: mapping the structure and functions of the nuclear pore complex.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Curr Opin Cell Biol. 2012 Feb;24(1):92-9. doi: 10.1016/j.ceb.2011.12.013. Epub 2012 Feb 8. Curr Opin Cell Biol. 2012. PMID: 22321828 Free PMC article. Review. - Nuclear pore complexes: guardians of the nuclear genome.
Capelson M, Doucet C, Hetzer MW. Capelson M, et al. Cold Spring Harb Symp Quant Biol. 2010;75:585-97. doi: 10.1101/sqb.2010.75.059. Epub 2011 Apr 18. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21502404 Free PMC article. Review. - Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions.
Raices M, D'Angelo MA. Raices M, et al. Nat Rev Mol Cell Biol. 2012 Nov;13(11):687-99. doi: 10.1038/nrm3461. Nat Rev Mol Cell Biol. 2012. PMID: 23090414 Review. - Architecture of the symmetric core of the nuclear pore.
Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, Hoelz A. Lin DH, et al. Science. 2016 Apr 15;352(6283):aaf1015. doi: 10.1126/science.aaf1015. Epub 2016 Apr 14. Science. 2016. PMID: 27081075 Free PMC article.
References
- Tran E. J., Wente S. R. (2006) Cell 125, 1041–1053 - PubMed
- Weis K. (2003) Cell 112, 441–451 - PubMed
- Schwartz T. U. (2005) Curr. Opin Struct. Biol. 15, 221–226 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous