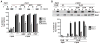Mechanistic analysis of a dynamin effector - PubMed (original) (raw)
Mechanistic analysis of a dynamin effector
Laura L Lackner et al. Science. 2009.
Abstract
Dynamin-related proteins (DRPs) can generate forces to remodel membranes. In cells, DRPs require additional proteins [DRP-associated proteins (DAPs)] to conduct their functions. To dissect the mechanistic role of a DAP, we used the yeast mitochondrial division machine as a model, which requires the DRP Dnm1, and two other proteins, Mdv1 and Fis1. Mdv1 played a postmitochondrial targeting role in division by specifically interacting and coassembling with the guanosine triphosphate-bound form of Dnm1. This regulated interaction nucleated and promoted the self-assembly of Dnm1 into helical structures, which drive membrane scission. The nucleation of DRP assembly probably represents a general regulatory strategy for this family of filament-forming proteins, similar to F-actin regulation.
Figures
Fig. 1
Mdv1 preferentially interacts with the GTP-bound form of Dnm1. (A) Dnm1 and the indicated Dnm1 mutants at 0.4 μM were incubated with liposomes in the absence and presence of various nucleotides. The association of the protein with liposomes was assessed by its ability to float with liposomes after equilibrium sucrose gradient centrifugation. Equivalent amounts of the top (T) and bottom (B) fractions of the gradients were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. The percentage of protein found in the top fraction is shown as the mean and SEM; n = 3 independent experiments. (B) Dnm1 and Mdv1, each at 0.4 μM, were incubated with liposomes in the absence and presence of various nucleotides and subjected to liposome floatation and analysis as described in (A). Data are shown as the mean and SEM; n = 3 independent experiments. *P ≤ 0.05.
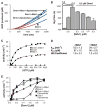
Fig. 2
Mdv1 acts to nucleate Dnm1 self-assembly. (A) A plot of Dnm1 GTPase activity in the absence and presence of Mdv1 and OMC liposomes versus time after the initiation of self-assembly. The lag time as determined by linear regression analysis is shown in red and the steady-state region of the curve is shown in blue. Dnm1 and Mdv1 were present at 0.5 and 0.4 μM, respectively. OMC liposomes were present at 0.05 mg/ml, and GTP was present at 500 μM. (B) Quantification of lag time in the presence of various concentrations of Mdv1. Dnm1 was present at 1.5, 0.8, or 0.5 μM, as indicated, and GTP was present at 500 μM. Data are shown as the mean and SEM; n = 3 independent experiments. (C andD) Steady-state kinetics of Dnm1 in the absence and presence of Mdv1. Dnm1 and Mdv1 were present at 0.5 and 0.4 μM, respectively. A representative kinetic experiment is shown in (C). The table in (D) shows kinetic parameters determined as described in methods (32). _k_cat, turnover number; _K_0.5, substrate concentration where velocity is one-half maximal. Data are shown as the mean and SEM; _n_= 3 independent experiments. (E) Steady-state GTP hydrolysis activity of Dnm1 at various Dnm1 concentrations in the absence and presence of 0.4 μM Mdv1 and 0.05 mg/ml OMC liposomes, as indicated. GTP was present at 500 μM. Data are shown as the mean and SEM;n = 3 independent experiments.
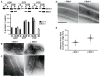
Fig. 3
Mdv1 stimulates Dnm1 self-assembly. (A) Velocity sedimentation of Dnm1 in the absence and presence of Mdv1. Dnm1 and Mdv1 were present at 0.2 μM. A representative Western blot is shown, and quantification data are shown as the mean and SEM; n = 3 independent experiments. (B and C) Analysis of GMPPCP-dependent Dnm1 self-assembly by negative-stain EM in the absence (left panels) and presence (right panels) of Mdv1. Different fields are shown in (B) and (C); the images in (C) were taken at a magnification 15,000 times higher than those in (B). Scale bars, 100 nm. (D) Negative-stain EM images of Dnm1-GMPPCP assembled on liposomes in the absence (left panel) and presence (right panel) of Mdv1. Scale bar, 100 nm. (E) Quantification of the distance from the inner edge of the lipid membrane to the outer edge of the assembled protein on Dnm1- and Dnm1-Mdv1 coated liposomes; _n_= 24 protein-coated liposomes. The mean and SEM are also shown.
Fig. 4
Mdv1 oligomerizes. (A) Hydrodynamic parameters of Mdv1 were determined by sucrose gradient and gel filtration analyses and calculated as described in methods (32). (B) Analysis of Mdv1 oligomerization by chemical cross-linking. SDS-PAGE Coomassie-stained gel is shown of Mdv1 samples without (lane 1) and with (lane 2) a 5-M excess of cross-link reagent.
Similar articles
- Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes.
Koirala S, Bui HT, Schubert HL, Eckert DM, Hill CP, Kay MS, Shaw JM. Koirala S, et al. J Cell Biol. 2010 Dec 13;191(6):1127-39. doi: 10.1083/jcb.201005046. J Cell Biol. 2010. PMID: 21149566 Free PMC article. - A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment.
Bui HT, Karren MA, Bhar D, Shaw JM. Bui HT, et al. J Cell Biol. 2012 Nov 12;199(4):613-22. doi: 10.1083/jcb.201207079. J Cell Biol. 2012. PMID: 23148233 Free PMC article. - Dnm1 forms spirals that are structurally tailored to fit mitochondria.
Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Ingerman E, et al. J Cell Biol. 2005 Sep 26;170(7):1021-7. doi: 10.1083/jcb.200506078. J Cell Biol. 2005. PMID: 16186251 Free PMC article. - The machines that divide and fuse mitochondria.
Hoppins S, Lackner L, Nunnari J. Hoppins S, et al. Annu Rev Biochem. 2007;76:751-80. doi: 10.1146/annurev.biochem.76.071905.090048. Annu Rev Biochem. 2007. PMID: 17362197 Review. - Molecular Basis of Mitochondrial and Peroxisomal Division Machineries.
Imoto Y, Itoh K, Fujiki Y. Imoto Y, et al. Int J Mol Sci. 2020 Jul 30;21(15):5452. doi: 10.3390/ijms21155452. Int J Mol Sci. 2020. PMID: 32751702 Free PMC article. Review.
Cited by
- When the Balance Tips: Dysregulation of Mitochondrial Dynamics as a Culprit in Disease.
Kyriakoudi S, Drousiotou A, Petrou PP. Kyriakoudi S, et al. Int J Mol Sci. 2021 Apr 28;22(9):4617. doi: 10.3390/ijms22094617. Int J Mol Sci. 2021. PMID: 33924849 Free PMC article. Review. - Mitochondrial network morphology: building an integrative, geometrical view.
Rafelski SM. Rafelski SM. BMC Biol. 2013 Jun 24;11:71. doi: 10.1186/1741-7007-11-71. BMC Biol. 2013. PMID: 23800141 Free PMC article. Review. - Cyclin C mediates stress-induced mitochondrial fission and apoptosis.
Wang K, Yan R, Cooper KF, Strich R. Wang K, et al. Mol Biol Cell. 2015 Mar 15;26(6):1030-43. doi: 10.1091/mbc.E14-08-1315. Epub 2015 Jan 21. Mol Biol Cell. 2015. PMID: 25609094 Free PMC article. - Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage.
Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Kageyama Y, et al. J Cell Biol. 2012 May 14;197(4):535-51. doi: 10.1083/jcb.201110034. Epub 2012 May 7. J Cell Biol. 2012. PMID: 22564413 Free PMC article. - Mitochondrial Dynamics in Stem Cells and Differentiation.
Seo BJ, Yoon SH, Do JT. Seo BJ, et al. Int J Mol Sci. 2018 Dec 5;19(12):3893. doi: 10.3390/ijms19123893. Int J Mol Sci. 2018. PMID: 30563106 Free PMC article. Review.
References
- Praefcke GJ, McMahon HT. Nat Rev Mol Cell Biol. 2004;5:133. - PubMed
- Hoppins S, Lackner L, Nunnari J. Annu Rev Biochem. 2007;76:751. - PubMed
- Hinshaw JE, Schmid SL. Nature. 1995;374:190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM097432/GM/NIGMS NIH HHS/United States
- R01 GM062942/GM/NIGMS NIH HHS/United States
- 1F32GM078749/GM/NIGMS NIH HHS/United States
- F32 GM078749/GM/NIGMS NIH HHS/United States
- R01GM062942/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases