SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation - PubMed (original) (raw)
. 2009 Sep 16;28(18):2748-62.
doi: 10.1038/emboj.2009.210. Epub 2009 Aug 13.
Yan Han, Yumei Wang, Xuxu Sun, Shan Yan, Edward T H Yeh, Yuying Chen, Hui Cang, Hui Li, Guiying Shi, Jinke Cheng, Xueming Tang, Jing Yi
Affiliations
- PMID: 19680224
- PMCID: PMC2750016
- DOI: 10.1038/emboj.2009.210
SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation
Chao Huang et al. EMBO J. 2009.
Abstract
The physiological function of Sentrin/SUMO-specific proteases (SENPs) remains largely unexplored, and little is known about the regulation of SENPs themselves. Here, we show that a modest increase of reactive oxygen species (ROS) regulates SENP3 stability and localization. We found that SENP3 is continuously degraded through the ubiquitin-proteasome pathway under basal condition and that ROS inhibit this degradation. Furthermore, ROS causes SENP3 to redistribute from the nucleoli to the nucleoplasm, allowing it to regulate nuclear events. The stabilization and redistribution of SENP3 correlate with an increase in the transcriptional activity of the hypoxia-inducing factor-1 (HIF-1) under mild oxidative stress. ROS-enhanced HIF-1 transactivation is blocked by SENP3 knockdown. The de-SUMOylating activity of SENP3 is required for ROS-induced increase of HIF-1 transactivation, but the true substrate of SENP3 is the co-activator of HIF-1 alpha, p300, rather than HIF-1 alpha itself. Removing SUMO2/3 from p300 enhances its binding to HIF-1 alpha. In vivo nude mouse xenografts overexpressing SENP3 are more angiogenic. Taken together, our results identify SENP3 as a redox sensor that regulates HIF-1 transcriptional activity under oxidative stress through the de-SUMOylation of p300.
Figures
Figure 1
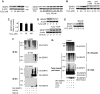
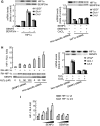
Mild oxidative stress induces a rapid stabilization of SENP3 protein. (A) HeLa cells were treated with the indicated concentrations of hydrogen peroxide (H2O2) for 1 h (left panel) or 100 μM H2O2 for the indicated time (right panel). SENP3 protein level was evaluated by immunoblotting (IB) using SENP3 antibody. β-actin was used as a loading control. (B) HeLa cells were pre-treated with 5 mM NAC for 4 h or 10 mM DTT for 1 h, before H2O2 was added to the medium, for an additional 1 h. SENP3 protein levels were evaluated by IB. (C) HeLa cells were treated with the indicated concentrations of H2O2 for 1 h. SENP3 mRNA levels were evaluated by real-time PCR and shown as folds of control. (D) SENP3 protein levels were evaluated by IB after HUVEC cells were treated with cycloheximide (CHX) for different times (upper panel), or pre-treated with H2O2 for 2 h before CHX was added and co-incubated for the indicated time (lower panel). β-actin was used as a loading control. (E) SENP3 protein levels were evaluated by IB after HUVEC cells were treated with H2O2 as indicated for 1 h with or without 30 μM MG132. (F) HEK293T cells were transfected with RH-SENP3 or/and HA-Ubiquitin (HA-Ub) for 36 h, and MG132 was then added for additional 10 h as indicated. Co-IP was carried out with RH antibody and IB was carried out as indicated. The blots with long time of exposure (long expo) and short time of exposure (short expo) are displayed to show the differences in quantity between ubiquitin-conjugated and non-conjugated SENP3, respectively. (G) HEK293T cells were transfected with RH-SENP3 for 36 h. MG132, H2O2 and DTT were added as indicated for an additional 1 h. RH-SENP3 was pulled down with Talon beads and IB was carried out using anti-ubiquitin and anti-RGS antibodies, respectively.
Figure 2
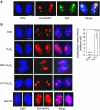
SENP3 redistributes between the nucleolus and the nucleoplasm on H2O2 exposure. (A) HeLa cells were transfected with RH-SENP3 for 36 h. Immunofluorescence was carried out with RH and B23 antibodies. B23 is a nucleolar marker. DAPI was used for counterstaining the nuclei. Scale bar=9 μm. (B) HeLa cells were transfected with RH-SENP3 for 36 h, and the cells were treated with H2O2 (100 μM) or MG132 (10 mM), respectively, for 1 h, or pretreated with 5 mM NAC for 4 h or 10 mM DTT for 1 h before H2O2 exposure for another 1 h. Immunofluorescence was carried out using RH antibody. DAPI was used for counterstaining the nuclei. Scale bar=9 μm. Bar charts showed the difference in the rhodamine intensity that reflected SENP3 quantity in the nucleoplasm in eighty cells with or without H2O2 treatment.
Figure 3
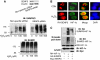
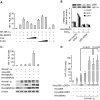
SENP3 participates in H2O2-induced changes in SUMO2/3 modification and can interact with HIF-1. (A) HeLa cells were transfected with non-specific siRNA and SENP3 siRNA for 72 h, and SENP3 expression was markedly knocked down (upper panel). The cells were then treated with various concentrations of H2O2 for 1 h before global protein SUMOylation was evaluated using the SUMO2/3 antibody (middle panel) and the SUMO1 antibody (bottom panel). (B) After being transfected with HA-HIF-1α and RH-SENP3 for 48 h, HeLa cells were exposed to 100 μM H2O2 for 1 h. Cell monolayers were fixed and immunofluorescence was carried out using anti-HIF-1α and anti-RH antibodies. Cell nuclei were stained with DAPI. Scale bar=9 μm. (C) After HeLa cells were transfected with RH-HIF-1α with or without HA-SUMO3 and Myc-SENP3 as indicated for 48 h, cells were lysed and RH-HIF-1α was pulled down using Ni beads. The pulled-down HIF-1α and cell lysates were analysed by IB as indicated.
Figure 4a
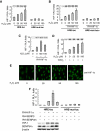
Mild oxidative stress enhances the transcriptional activity of HIF-1 through SENP3, but this action is not attributed to the de-SUMOylation of HIF-1α by SENP3. (A–D) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE or HRE mutant, and Renilla, with or without RH-HIF-1α. At 40 h post-transfection, cells were exposed to the indicated concentrations of H2O2 and CoCl2, as described in Materials and methods section, before cells were lysed, and the relative luciferase activity (RLA) for HIF-1 was assayed. NAC, when used, was pre-incubated with cells for 4 h before H2O2 was added to the medium. The cell lysates of one of the representative experiments were subjected to IB with anti-HIF-1α antibody. The results of the RLA are given as the mean±s.d. of three independent experiments, and samples were duplicated in each experiment. (E) After HeLa cells were exposed to the indicated concentration of H2O2 for 1 h, cell monolayers were fixed and immunofluorescence was carried out using anti-HIF-1α antibody. Scale bar=10 μm. (F) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE or HRE mutant, and Renilla, along with RH-HIF-1α, RH-SENP3 or RH-SENP3 mutant were additionally co-transfected as indicated. At 48 h post-transfection, cells were lysed and HIF-1 RLA was assayed.
Figure 4b
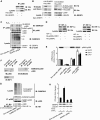
(G) HeLa cells were co-transfected with the constructs for RH-HIF-1α, with or without RH-SENP3 or RH-SENP3 mutant as indicated (left panel), or with shRNA to knock down endogenous HIF-1α (right panel). At 48 h after transfection of SENP3 constructs, or 42 h post-transfection of HIF-1α shRNA, plus treatment with 150 μM CoCl2 for 6 h, cells were collected for real-time PCR to determine the expression of three major HIF-1 target genes as indicated. The levels of mRNA were shown as folds of control. (H) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE and Renilla, and RH-HIF-1α, together with SENP3 siRNA or non-specific siRNA as indicated. At 72 h post-transfection, cells were treated with the indicated concentrations of H2O2 before they were lysed and HIF-1 RLA was assayed (left panel). HeLa cells were transfected with siRNA to knock down endogenous SENP3 (right panel). At 66 h after transfection of siRNA, plus treatment with 150 μM CoCl2 for 6 h, cells were collected for real-time PCR to determine the expression of the HIF-1 target genes as indicated. (I) HeLa cells were transfected with the constructs of luciferase reporters for HRE and Renilla, along with wild type (wt) HIF-1α, or SUMOylation sites-mutated (sm) HIF-1α (K391, 477R) with or without increasing amounts of RH-SENP3 or RH-SENP3 mutant as indicated. At 48 h post-transfection, the cells were lysed and RLA was assayed.
Figure 5
The effect of SENP3 on the enhancement of HIF-1 transcriptional activity depends on p300 and its de-SUMOylation. (A) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE and Renilla, RH-HIF-1α and RH-SENP3, and increasing amounts of E1A or E1A mutant as indicated. At 48 h post-transfection, the cells were lysed and HIF-1 RLA was assayed. (B) HeLa cells were transfected with siRNA to knock down endogenous p300. At 66 h post-transfection of siRNA, cells were incubated with 150 μM CoCl2 for 6 h and then collected for real-time PCR. (C) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE and Renilla, RH-HIF-1α and RH-SENP3 or its mutant. His-p300 was additionally co-transfected as indicated. At 48 h post-transfection, the cells were lysed and HIF-1 RLA was assayed. (D) HeLa cells were co-transfected with the constructs of luciferase reporters for HRE and Renilla, RH-HIF-1α and RH-SENP3. His-p300 or His-p300ΔCRD1 were additionally co-transfected as indicated. p300ΔCRD1 is a p300 truncate that lacks the domain containing two sites for SUMOylation and is thus unable to be SUMOylated. At 48 h post-transfection, the cells were lysed and HIF-1 RLA was assayed. The results showed that overexpression of p300ΔCRD1 (the sixth bar, left to right) alone could promote the HIF-1 transactivation to an extent much higher than what its full-length counterpart did (compare the sixth bar with the third, with statistically significant difference), and comparable to the synergistic effect of SENP and full-length p300 (compare the sixth bar with the fifth). SENP3 could not further promote the enhancing effect of p300ΔCRD1 (compare the sixth bar with the seventh, with statistically insignificant difference).
Figure 6
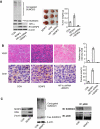
p300 is a direct substrate of SENP3 and de-SUMOylation of p300 potentiates its interaction with HIF-1. (A) HeLa cells were transfected with the constructs for His-p300 alone or RH-SENP3 as indicated. At 48 h post-transfection, the cells were lysed and co-IP was carried out with anti-p300 antibody. Bound proteins and cell lysates were analysed by IB as indicated. (B) HeLa cells were co-transfected with His-p300, and HA-SUMO2 or HA-SUMO3, with or without Myc-SENP3 as indicated. At 48 h post-transfection, the cells were lysed and a pull-down assay was performed using Ni-beads. Bound proteins and cell lysates were analysed by IB as indicated. (C) HEK293T cells were transfected with non-specific siRNA or SENP3 siRNA for 72 h to deplete endogenous SENP3. The cells were treated with 100 μM H2O2 for 1 h before they were lysed and p300 was pulled down using p300 antibody. Bound proteins and cell lysates were analysed by IB as indicated. (D) HeLa cells were transfected with HIF-1α and His-p300, and His-p300ΔCRD1 (left panel) or RH-SENP3 (right panel) as indicated. At 48 h post-transfection, the cells were lysed and IP was carried out using anti-HIF-1α and IB was carried out as indicated. (E) HeLa cells were co-transfected with siRNA specific to endogenous p300 and/or wild-type or truncated p300 constructs as indicated for 72 h. The expressions of three major HIF-1 target genes were determined by real-time PCR. (F) HeLa cells were transfected with HA-SUMO3 for 48 h. Endogenous p300 that bound with HIF-1α in HeLa cells was immunoprecipitated using anti-HIF-1α antibody, whereas p300 in the rest portion, that is, in the supernatant was immunoprecipitated using anti-p300 antibody. Samples were then detected by anti-p300, anti-SUMO2/3 and anti-HIF-1α antibodies, respectively. (G) HeLa cells were transfected with non-specific siRNA or siRNAs for SUMO2 and SUMO3. At 72 h post-transfection, the cells were lysed and p300 was immunoprecipitated using anti-p300 antibody. The efficiency of SUMO2/3 silencing was evaluated in whole-cell lysates with anti-SUMO2/3 antibody. The SUMOylation status of p300 was determined using anti-SUMO2/3 and anti-p300 antibodies. The whole-cell lysates were then incubated with _in vitro_-translated RH-tagged HIF-1α for 1 h. IP was carried out using anti-RH antibody, and IB was carried out using anti-p300 and anti-RH antibodies. (H) Endogenous SENP3 was knocked down by siRNA and cells were exposed to H2O2 for 1 h. p300 that was recruited to HRE DNA was immunoprecipitated using anti-p300 antibody. The bound HRE in the precipitates were then quantitatively analysed by real-time PCR.
Figure 7
Overexpression of SENP3 promotes in vivo angiogenesis in tumour xenografts. (A) HeLa cells were stably transfected with empty vector+non-specific shRNA (as a control group), SENP3+non-specific shRNA (as SENP3 group) or SENP3+HIF-1α shRNA (as SENP3+HIF-1α shRNA group). Cell colonies were examined for specificity and efficiency of protein expressions and SUMO2/3 modification pattern (left panel). The cells were injected subcutaneously into the left flank of 4-week-old BALB/c-nu-nu mice. After 3 weeks, the mice were sacrificed and the tumour xenografts were dissected (middle panel). The bar chart showed mean±s.d. of the tumour weights (right panel). (B) In situ hybridization for human VEGF mRNA (left, upper panel, blue-purple signal), and immunohistochemistry for mouse CD31 (left, lower panel, yellow-brown signal) in tumor tissues. The bar charts showed the relative hybridization signal intensity (right, upper panel) and the areas positive for CD31 (right, lower panel) in tumour tissues. Scale bar=10 μM. (C) The mixed tumour tissues were homogenized. The enforced-expressed SENP3 (left panel) and SUMO2/3 modification pattern (middle panel) were examined by IB. P300 was immunoprecipitated using anti-p300 antibody, and examined for SUMOylation status by IB as indicated (right panel).
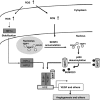
Figure 8
The hypothetical illustration for a role of SENP3 and de-SUMO2/3 of p300 in ROS-enhanced HIF-1 transactivation.
Similar articles
- The biphasic redox sensing of SENP3 accounts for the HIF-1 transcriptional activity shift by oxidative stress.
Wang Y, Yang J, Yang K, Cang H, Huang XZ, Li H, Yi J. Wang Y, et al. Acta Pharmacol Sin. 2012 Jul;33(7):953-63. doi: 10.1038/aps.2012.40. Epub 2012 Jun 11. Acta Pharmacol Sin. 2012. PMID: 22684029 Free PMC article. - Dynamic regulation of HIF1Α stability by SUMO2/3 and SENP3 in the human placenta.
Bhattacharjee J, Alahari S, Sallais J, Tagliaferro A, Post M, Caniggia I. Bhattacharjee J, et al. Placenta. 2016 Apr;40:8-17. doi: 10.1016/j.placenta.2016.02.002. Epub 2016 Feb 6. Placenta. 2016. PMID: 27016777 - Overexpression of SENP3 in oral squamous cell carcinoma and its association with differentiation.
Sun Z, Hu S, Luo Q, Ye D, Hu D, Chen F. Sun Z, et al. Oncol Rep. 2013 May;29(5):1701-6. doi: 10.3892/or.2013.2318. Epub 2013 Mar 1. Oncol Rep. 2013. PMID: 23467634 Free PMC article. - The Critical Roles of the SUMO-Specific Protease SENP3 in Human Diseases and Clinical Implications.
Long X, Zhao B, Lu W, Chen X, Yang X, Huang J, Zhang Y, An S, Qin Y, Xing Z, Shen Y, Wu H, Qi Y. Long X, et al. Front Physiol. 2020 Oct 30;11:558220. doi: 10.3389/fphys.2020.558220. eCollection 2020. Front Physiol. 2020. PMID: 33192553 Free PMC article. Review. - Cellular redox status regulates hypoxia inducible factor-1 activity. Role in tumour development.
Martínez-Sánchez G, Giuliani A. Martínez-Sánchez G, et al. J Exp Clin Cancer Res. 2007 Mar;26(1):39-50. J Exp Clin Cancer Res. 2007. PMID: 17550131 Review.
Cited by
- Identification and characterization of a new chemotype of noncovalent SENP inhibitors.
Madu IG, Namanja AT, Su Y, Wong S, Li YJ, Chen Y. Madu IG, et al. ACS Chem Biol. 2013 Jul 19;8(7):1435-41. doi: 10.1021/cb400177q. Epub 2013 May 1. ACS Chem Biol. 2013. PMID: 23614497 Free PMC article. - SENP1 participates in the dynamic regulation of Elk-1 SUMOylation.
Witty J, Aguilar-Martinez E, Sharrocks AD. Witty J, et al. Biochem J. 2010 May 13;428(2):247-54. doi: 10.1042/BJ20091948. Biochem J. 2010. PMID: 20337593 Free PMC article. - Inhibition of fatty acid oxidation enables heart regeneration in adult mice.
Li X, Wu F, Günther S, Looso M, Kuenne C, Zhang T, Wiesnet M, Klatt S, Zukunft S, Fleming I, Poschet G, Wietelmann A, Atzberger A, Potente M, Yuan X, Braun T. Li X, et al. Nature. 2023 Oct;622(7983):619-626. doi: 10.1038/s41586-023-06585-5. Epub 2023 Sep 27. Nature. 2023. PMID: 37758950 Free PMC article. - The Krüppel-like factor 15 as a molecular link between myogenic factors and a chromosome 4q transcriptional enhancer implicated in facioscapulohumeral dystrophy.
Dmitriev P, Petrov A, Ansseau E, Stankevicins L, Charron S, Kim E, Bos TJ, Robert T, Turki A, Coppée F, Belayew A, Lazar V, Carnac G, Laoudj D, Lipinski M, Vassetzky YS. Dmitriev P, et al. J Biol Chem. 2011 Dec 30;286(52):44620-31. doi: 10.1074/jbc.M111.254052. Epub 2011 Sep 21. J Biol Chem. 2011. PMID: 21937448 Free PMC article. - SUMO3 modification accelerates the aggregation of ALS-linked SOD1 mutants.
Niikura T, Kita Y, Abe Y. Niikura T, et al. PLoS One. 2014 Jun 27;9(6):e101080. doi: 10.1371/journal.pone.0101080. eCollection 2014. PLoS One. 2014. PMID: 24971881 Free PMC article.
References
- Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW (2004) Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun 324: 394–400 - PubMed
- Biswas S, Gupta MK, Chattopadhyay D, Mukhopadhyay CK (2007) Insulin-induced activation of hypoxia-inducible factor-1 requires generation of reactive oxygen species by NADPH oxidase. Am J Physiol Heart Circ Physiol 292: H758–H766 - PubMed
- Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21: 349–357 - PubMed
- Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS (2005) Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab 1: 409–414 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous