Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier - PubMed (original) (raw)
Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier
Bracha Naim et al. EMBO J. 2009.
Abstract
To fulfil their function, nuclear pore complexes (NPCs) must discriminate between inert proteins and nuclear transport receptors (NTRs), admitting only the latter. This specific permeation is thought to depend on interactions between hydrophobic patches on NTRs and phenylalanine-glycine (FG) or related repeats that line the NPC. Here, we tested this premise directly by conjugating different hydrophobic amino-acid analogues to the surface of an inert protein and examining its ability to cross NPCs unassisted by NTRs. Conjugation of as few as four hydrophobic moieties was sufficient to enable passage of the protein through NPCs. Transport of the modified protein proceeded with rates comparable to those measured for the innate protein when bound to an NTR and was relatively insensitive both to the nature and density of the amino acids used to confer hydrophobicity. The latter observation suggests a non-specific, small, and plant interaction network between cargo and FG repeats.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
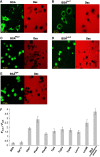
Nuclear entry of BSA molecules modified by amino-acid side-chain analogues on their surface. The modifier used in each panel is denoted in superscript by the three-letter code of the corresponding amino-acid side chain it carries and the number indicates the degree of modification. (A–E) Confocal images of digitonin-permeabilized, nocodazole-treated HeLa cells 30 min after the introduction of a transport mixture containing innate or modified BSA (green) and a 66-kDa dextran (red) probe serving as an NE integrity marker. (F) Quantitative analysis of nuclear entry. Shown is the ratio between (background-corrected) nuclear and cytoplasmic mean fluorescence intensity (_F_nuc/_F_cyt) measured 30 min after introduction of innate BSA, BSA molecules modified by different amino-acid analogues, and BSA linked to SV40 large T-antigen NLS peptides (in the presence of cytosolic extract and an energy regenerating system). Data are presented as the mean±2 s.e.m. of determinations involving 20–30 cells for each column.
Figure 2
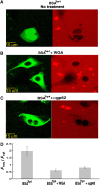
Inhibition of modified BSA transport by NPC blockers. (A–C) Confocal images of HeLa cells 30 min after the introduction of BSATrp4 either alone (A) or together with WGA (B) or αgp62 (C). The latter two are known blockers of NPC-mediated transport. Cells were permeabilized twice with the pore forming protein SLO, which is used for reversible permeabilization of the plasma membrane (Walev et al, 2001) (see Materials and methods) with the transport substrate introduced during the second permeabilization. (D) Quantitative analysis of nuclear entry. Shown is the ratio between (background-corrected) nuclear and cytoplasmic mean fluorescence intensity (_F_nuc/_F_cyt) measured 30 min after the second SLO permeabilization in which BSATrp4 was introduced to the cells. Data are presented as the mean±2 s.e.m. of determinations involving 6–12 cells for each column. Note that nuclear entry of the modified BSA in the absence of the blockers was as effective as that observed in digitonin-permeabilized cells.
Figure 3
Nuclear accumulation versus time traces of modified BSA. Shown are traces of BSA modified by (A) phenylalanine, (B) tryptophan, or (C) leucine analogues to varying degrees. Each data point represents the average obtained from multiple traces (see Figure 4C, for exact numbers) that were normalized before averaging. For convenience of presentation, error bars were not included (but see Figures 4A and 4C). The fits shown (solid lines) are provided only as guidance to the eyes.
Figure 4
Nuclear import kinetics of BSA modified by either hydrophobic amino acids or by NLSs and of an NTR. (A) A plot of _t_1/2 values measured for all BSA derivatives used in this study (blue circles) versus N—the estimated number of hydrophobic moieties attached to the protein. With our experimental setup and timeframe of the measurements, we could not accurately determine the nuclear entry rate of unmodified BSA because it was too low. It has been estimated that BSA traffics to the nucleus at least 600 times slower than transportin (Ribbeck and Gorlich, 2001). Combining this with our data, we estimate that native BSA enters the nucleus with a _t_1/2 of about 50 000 s, under the experimental conditions we used. This latter value is marked in the graph by a black square (note the break in the y axis). (B) Comparison between nuclear import kinetics of BSALeu66 and BSA derivatized by NLSs, which serve as substrates for the transport receptor complex importin α/β. In these experiments, the cells were supplemented by cytoplasmic extract and an energy regenerating system. (Inset) Nuclear import kinetics of BSALeu66 as compared with that of the (unloaded) nuclear import receptor transportin (fused to GST), both measured in unsupplemented cells. Data describing the nuclear import of the transport receptor were taken from Naim et al (2007). (C) _t_1/2 values derived for nuclear transport of BSA derivatives used in this study. The numbers in parenthesis denote the number of cells used in the analysis.
Similar articles
- Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes.
Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. Labokha AA, et al. EMBO J. 2013 Jan 23;32(2):204-18. doi: 10.1038/emboj.2012.302. Epub 2012 Nov 30. EMBO J. 2013. PMID: 23202855 Free PMC article. - Large cargo transport by nuclear pores: implications for the spatial organization of FG-nucleoporins.
Tu LC, Fu G, Zilman A, Musser SM. Tu LC, et al. EMBO J. 2013 Dec 11;32(24):3220-30. doi: 10.1038/emboj.2013.239. Epub 2013 Nov 8. EMBO J. 2013. PMID: 24213245 Free PMC article. - Emergence of selectivity and specificity in a coarse-grained model of the nuclear pore complex with sequence-agnostic FG-Nups.
Patel MK, Chakrabarti B, Panwar AS. Patel MK, et al. Phys Chem Chem Phys. 2023 Dec 13;25(48):32824-32836. doi: 10.1039/d3cp03746k. Phys Chem Chem Phys. 2023. PMID: 38018404 - The selective permeability barrier in the nuclear pore complex.
Li C, Goryaynov A, Yang W. Li C, et al. Nucleus. 2016 Sep 2;7(5):430-446. doi: 10.1080/19491034.2016.1238997. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27673359 Free PMC article. Review. - Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality.
Peters R. Peters R. Traffic. 2005 May;6(5):421-7. doi: 10.1111/j.1600-0854.2005.00287.x. Traffic. 2005. PMID: 15813752 Review.
Cited by
- Characterization of the transport signals that mediate the nucleocytoplasmic traffic of low risk HPV11 E7.
McKee CH, Onder Z, Ashok A, Cardoso R, Moroianu J. McKee CH, et al. Virology. 2013 Aug 15;443(1):113-22. doi: 10.1016/j.virol.2013.04.031. Epub 2013 May 29. Virology. 2013. PMID: 23725695 Free PMC article. - The mechanistic role of alpha-synuclein in the nucleus: impaired nuclear function caused by familial Parkinson's disease SNCA mutations.
Chen V, Moncalvo M, Tringali D, Tagliafierro L, Shriskanda A, Ilich E, Dong W, Kantor B, Chiba-Falek O. Chen V, et al. Hum Mol Genet. 2020 Nov 4;29(18):3107-3121. doi: 10.1093/hmg/ddaa183. Hum Mol Genet. 2020. PMID: 32954426 Free PMC article. - Mutant libraries reveal negative design shielding proteins from supramolecular self-assembly and relocalization in cells.
Garcia Seisdedos H, Levin T, Shapira G, Freud S, Levy ED. Garcia Seisdedos H, et al. Proc Natl Acad Sci U S A. 2022 Feb 1;119(5):e2101117119. doi: 10.1073/pnas.2101117119. Proc Natl Acad Sci U S A. 2022. PMID: 35078932 Free PMC article. - Energetics of Transport through the Nuclear Pore Complex.
Ghavami A, van der Giessen E, Onck PR. Ghavami A, et al. PLoS One. 2016 Feb 19;11(2):e0148876. doi: 10.1371/journal.pone.0148876. eCollection 2016. PLoS One. 2016. PMID: 26894898 Free PMC article. - Molecular determinants of large cargo transport into the nucleus.
Paci G, Zheng T, Caria J, Zilman A, Lemke EA. Paci G, et al. Elife. 2020 Jul 21;9:e55963. doi: 10.7554/eLife.55963. Elife. 2020. PMID: 32692309 Free PMC article.
References
- Bayliss R, Littlewood T, Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102: 99–108 - PubMed
- Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J Biol Chem 277: 50597–50606 - PubMed
- Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Gorlich D, Stewart M (1999) Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 293: 579–593 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources