Characterisation of the passive permeability barrier of nuclear pore complexes - PubMed (original) (raw)
Characterisation of the passive permeability barrier of nuclear pore complexes
Dagmar Mohr et al. EMBO J. 2009.
Abstract
Nuclear pore complexes (NPCs) restrict uncontrolled nucleocytoplasmic fluxes of inert macromolecules but permit facilitated translocation of nuclear transport receptors and their cargo complexes. We probed the passive barrier of NPCs and observed sieve-like properties with a dominating mesh or channel radius of 2.6 nm, which is narrower than proposed earlier. A small fraction of diffusion channels has a wider opening, explaining the very slow passage of larger molecules. The observed dominant passive diameter approximates the distance of adjacent hydrophobic clusters of FG repeats, supporting the model that the barrier is made of FG repeat domains cross-linked with a spacing of an FG repeat unit length. Wheat germ agglutinin and the dominant-negative importin beta(45-462) fragment were previously regarded as selective inhibitors of facilitated NPC passage. We now observed that they do not distinguish between the passive and the facilitated mode. Instead, their inhibitory effect correlates with the size of the NPC-passing molecule. They have little effect on small species, inhibit the passage of green fluorescent protein-sized objects >10-fold and virtually block the translocation of larger ones. This suggests that passive and facilitated NPC passage proceed through one and the same permeability barrier.
Figures
Figure 1
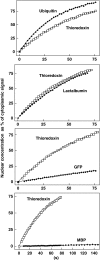
Time course of diffusion for various inert substrates into nuclei. Alexa488-labelled ubiquitin, lactalbumin or MBP as well as GFP were each pre-mixed with Alexa568-labelled thioredoxin. Entry of the substrate pairs into nuclei of permeabilised cells was followed by confocal laser scanning microscopy in separate fluorescence channels. Four time points each are shown. For quantitation see Figure 2. Post-experimental controls show nuclear exclusion of Alexa647-MBP and active import of IBB-GFP (see main text as well as Materials and methods).
Figure 2
Quantitation of passive entry into nuclei. Graphs show evaluations of the time series from Figure 1. Nuclear substrate concentrations are plotted versus time. Each graph shows the indicated diffusion substrate (⧫) and thioredoxin as a control (□). Data points were fitted to c(t)=a+k_·_t for MBP and GFP, and to c(t)=c_max·(1−e−_k_·_t) for all other proteins. Averages of analysed cells are shown. Nuclear entry rates depend on the size of these inert substrates. Although nuclear entry of ubiquitin, lactalbumin and thioredoxin occurred rapidly, influx of GFP (27 kDa) was slow and that of MBP (40 kDa) was very slow.
Figure 3
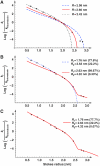
Estimation of passive pore radii from passage rates. Graph shows a plot of Stokes radii (r i) versus the natural logarithm of k i/_k_thioredoxin, the ratios of nuclear entry rates. The following probes were included: a fluorescein maleimide-cysteine adduct (0.67 nm Stokes radius), a labelled 11 aa peptide (0.9 nm), insulin (1.19 nm), aprotinin (1.48 nm), profilin 1 (1.65 nm), a z-domain from Protein A (1.71 nm), thioredoxin (1.97 nm), lactalbumin (2.07 nm), GFP (2.42 nm), the phosphate-binding protein PBP (2.75 nm) and the maltose-binding protein MBP (2.85 nm). The data were parameter-fitted to different models for passive passage through NPCs (see equations (5) and (7)). A faithful model, consistent with all data points, could only be obtained with the assumption of heterogeneity in channel width. A smaller residuum indicates a better fit of the data (see below). For details see main text and Materials and methods. (A) ‘Uniformly sized channel model', as detailed in equation (5). The best fit was obtained for _R_=2.66 nm with a residuum of 3.58 (red curve). This fit, however, does not explain the very slow NPC passage of PBP and MBP. The panel also shows a simulated blue dashed curve with _R_=2.98 nm (residuum=5.87), which predicts best the flux ratio for MBP and thioredoxin. The gray dotted curve was obtained for _R_=2.42 (residuum=4.46); it predicts the flux ratios for small probes best. (B) ‘Two radii model' (with _n_=2 according to equation (7), Materials and methods). The radius combination of 1.76 and 2.63 nm gave a near-perfect fit for small and medium-sized probes (blue dotted line, residuum=2.75), whereas the combination 2.63 and 4.32 nm gave an excellent fit for medium sized and larger probes (red line, best global fit, residuum=1.51). (C) ‘Three radii model' (with _n_=3, equation (7)), which gave an excellent fit for small, medium sized and large probes (residuum=0.39).
Figure 4
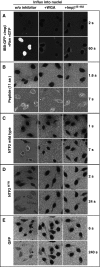
Effects of WGA and Impβ45-462 on facilitated and passive NPC passage. Permeabilised cells were either left untreated (left row) or pre-incubated with 0.1 mg/ml wheat germ agglutinin (middle row) or with 3 μM of the dominant-negative importin β45-462 fragment (right row). Thereafter, indicated influx experiments were performed. Import of IBB-GFP was performed in the presence of importin β, Ran and a GTP-regenerating system. All other import mixtures comprised only the fluorescent proteins. WGA and Impβ45-462 inhibited both, facilitated NPC passage (of IBB-GFP·importin β or NTF2) and passive nuclear entry of e.g. GFP. The inhibitory effects were strongest towards the large (132 kDa) IBB-GFP·importin β complex, strong towards GFP-sized objects (NTF2, NTF2W7R, GFP), and non-significant towards the smallest substrate, an 11 amino acid long peptide. For quantitation, see Figure 5.
Figure 5
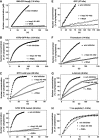
Quantitative analysis of the effects imposed by WGA and Impβ45-462. Graphs compare the entry kinetics of a given substrate into untreated nuclei (▪) with entry into nuclei pre-incubated with either WGA (▿) or Impβ45-462 (○). See also Figure 4 and Table II.
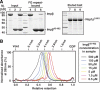
Figure 6
Impβ45-462 multimerises and binds FG repeats more strongly than full-length importin β. (A) Nickel-Sepharose beads coated with the His-tagged FG/FxFG repeat domain of Nsp1p were incubated with an excess of untagged Impβ (lanes 1, 4, 7), untagged Impβ45-452 (lanes 2, 5, 8) or a mixture of both proteins at physiological salt conditions. Bound NTRs and bait proteins were sequentially eluted and analysed by SDS–PAGE and Coomassie staining. Loaded material corresponds to 0.75% of input material or 3% of the eluted material. In comparison to the single incubations, binding of Impβ was drastically diminished in the presence of Impβ45-452, whereas binding of the Impβ mutant itself was virtually unaffected. This indicates that Impβ45-452 has an increased affinity for FG repeat domains. (B) Impβ45-452 was mixed with plasmid DNA and GDP as markers for void volume and total volume, respectively, diluted to the indicated concentrations and subjected to analytical gel filtration. Absorbance profiles acquired at 280 nm were normalised to the maximal optical density. Note that the observed peak positions and shapes greatly depend on the protein concentration of the applied sample. Higher order oligomeric forms are observed at higher protein concentrations. The gradual decrease of apparent molecular weight with higher dilutions indicates dynamic association/dissociation equilibria between multiple association states.
Similar articles
- Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes.
Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. Labokha AA, et al. EMBO J. 2013 Jan 23;32(2):204-18. doi: 10.1038/emboj.2012.302. Epub 2012 Nov 30. EMBO J. 2013. PMID: 23202855 Free PMC article. - The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion.
Ribbeck K, Görlich D. Ribbeck K, et al. EMBO J. 2002 Jun 3;21(11):2664-71. doi: 10.1093/emboj/21.11.2664. EMBO J. 2002. PMID: 12032079 Free PMC article. - FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties.
Frey S, Görlich D. Frey S, et al. EMBO J. 2009 Sep 2;28(17):2554-67. doi: 10.1038/emboj.2009.199. Epub 2009 Aug 13. EMBO J. 2009. PMID: 19680227 Free PMC article. - The selective permeability barrier in the nuclear pore complex.
Li C, Goryaynov A, Yang W. Li C, et al. Nucleus. 2016 Sep 2;7(5):430-446. doi: 10.1080/19491034.2016.1238997. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27673359 Free PMC article. Review. - Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease.
Jamali T, Jamali Y, Mehrbod M, Mofrad MR. Jamali T, et al. Int Rev Cell Mol Biol. 2011;287:233-86. doi: 10.1016/B978-0-12-386043-9.00006-2. Int Rev Cell Mol Biol. 2011. PMID: 21414590 Review.
Cited by
- Rapid Brownian Motion Primes Ultrafast Reconstruction of Intrinsically Disordered Phe-Gly Repeats Inside the Nuclear Pore Complex.
Moussavi-Baygi R, Mofrad MR. Moussavi-Baygi R, et al. Sci Rep. 2016 Jul 29;6:29991. doi: 10.1038/srep29991. Sci Rep. 2016. PMID: 27470900 Free PMC article. - Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier.
Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. Lin YC, et al. Nat Chem Biol. 2013 Jul;9(7):437-43. doi: 10.1038/nchembio.1252. Epub 2013 May 12. Nat Chem Biol. 2013. PMID: 23666116 Free PMC article. - The mechanistic role of alpha-synuclein in the nucleus: impaired nuclear function caused by familial Parkinson's disease SNCA mutations.
Chen V, Moncalvo M, Tringali D, Tagliafierro L, Shriskanda A, Ilich E, Dong W, Kantor B, Chiba-Falek O. Chen V, et al. Hum Mol Genet. 2020 Nov 4;29(18):3107-3121. doi: 10.1093/hmg/ddaa183. Hum Mol Genet. 2020. PMID: 32954426 Free PMC article. - Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia.
Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, Khokha MK. Del Viso F, et al. Dev Cell. 2016 Sep 12;38(5):478-92. doi: 10.1016/j.devcel.2016.08.002. Epub 2016 Sep 1. Dev Cell. 2016. PMID: 27593162 Free PMC article. - Simple rules for passive diffusion through the nuclear pore complex.
Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. Timney BL, et al. J Cell Biol. 2016 Oct 10;215(1):57-76. doi: 10.1083/jcb.201601004. Epub 2016 Oct 3. J Cell Biol. 2016. PMID: 27697925 Free PMC article.
References
- Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Görlich D, Stewart M (1999) Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 293: 579–593 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases