Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases - PubMed (original) (raw)
doi: 10.1038/nbt.1562. Epub 2009 Aug 13.
Frank Soldner, Caroline Beard, Qing Gao, Maisam Mitalipova, Russell C DeKelver, George E Katibah, Ranier Amora, Elizabeth A Boydston, Bryan Zeitler, Xiangdong Meng, Jeffrey C Miller, Lei Zhang, Edward J Rebar, Philip D Gregory, Fyodor D Urnov, Rudolf Jaenisch
Affiliations
- PMID: 19680244
- PMCID: PMC4142824
- DOI: 10.1038/nbt.1562
Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases
Dirk Hockemeyer et al. Nat Biotechnol. 2009 Sep.
Abstract
Realizing the full potential of human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) requires efficient methods for genetic modification. However, techniques to generate cell type-specific lineage reporters, as well as reliable tools to disrupt, repair or overexpress genes by gene targeting, are inefficient at best and thus are not routinely used. Here we report the highly efficient targeting of three genes in human pluripotent cells using zinc-finger nuclease (ZFN)-mediated genome editing. First, using ZFNs specific for the OCT4 (POU5F1) locus, we generated OCT4-eGFP reporter cells to monitor the pluripotent state of hESCs. Second, we inserted a transgene into the AAVS1 locus to generate a robust drug-inducible overexpression system in hESCs. Finally, we targeted the PITX3 gene, demonstrating that ZFNs can be used to generate reporter cells by targeting non-expressed genes in hESCs and hiPSCs.
Figures
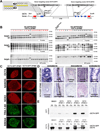
Figure 1. Targeting of OCT4 in hESCs using ZFNs
A. Schematic overview depicting the targeting strategy for the OCT4 locus. Probes used for Southern blot analysis are shown as red boxes, exons of the OCT4 locus are shown as blue boxes and arrows indicate the genomic site cut by the respective ZFN pair. Donor plasmids corresponding to the cleavage location of the three ZFN pairs carried 5’ and 3’ homology regions covering roughly 700 bp of the OCT4 sequence flanking the respective DSB target site are shown above; SA-GFP: splice acceptor eGFP sequence, 2A: self-cleaving peptide sequence, PURO: puromycin resistance gene, polyA: polyadenylation sequence. Inset in the upper left depicts a cartoon of two ZFNs binding at a specific genomic site (yellow) leading to the dimerization of the FokI nuclease domains. B. Southern blot analysis of BG01 cells targeted with the indicated ZFN pairs using the corresponding donor plasmids. Genomic DNA was digested either with EcoRI and hybridized with the external 3’-probe or digested with SacI and hybridized with the external 5’-probe or internal eGFP probe. Correctly targeted clones without additional integrations are indicated in red. C. Immunofluorescence staining of BG01 cells targeted with the indicated ZFN pairs using the corresponding donor plasmids. Cells were stained for the pluripotency markers OCT4, NANOG, SOX2, Tra-1–60 and SSEA4. D. Hematoxylin and eosin staining of teratoma sections generated from BG01 cells targeted with the indicated ZFN pairs and the corresponding donor plasmids. E. Western blot analysis for the expression of OCT4 and eGFP in BGO1 wild type cells and BG01 cells targeted with the indicated ZFN pairs using the corresponding donor plasmids. Cell extracts were derived from either undifferentiated cells (ES) or in vitro differentiated fibroblast-like cells (Fib.)
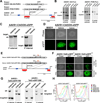
Figure 2. Targeting of the AAVS1 locus using ZFNs
A. Schematic overview depicting the targeting strategy for the PPP1R12C gene in the AAVS1 locus. Probes used for Southern blot analysis are shown as red boxes, the first 3 exons of PPP1R12C gene are shown as blue boxes; the arrow indicates the genomic site cut by the AAVS1-ZFNs. Donor plasmids used to target the locus are shown above; SA-Puro: splice acceptor sequence followed by a 2A self-cleaving peptide sequence and the puromycin resistance gene, pA: polyadenylation sequence, PGK: human phophoglycerol kinase promotor, PURO: puromycin resistance gene. B. Southern blot analysis of BG01 cells targeted with the indicated donor plasmids using the AAVS1 ZFNs. Genomic DNA was digested with SphI and hybridized with the 32P-labeled external 3’-probe or with the internal 5’-probe. Fragment sizes are: PGK-Puro: 5’-probe: wt=6.5 kb, targeted=4.2 kb; 3’-probe: wt=6.5 kb, targeted=3.7 kb. SA-Puro: 5’-probe: wt=6.5 kb, targeted=3.8 kb; 3’-probe: wt=6.5 kb, targeted=3.7 kb. C. Southern blot analysis of BG01 cells targeted with an AAVS1 donor plasmid containing a CAGGs driven eGFP cassette using the AAVS1 ZFNs. Genomic DNA was digested with SphI and hybridized with the 32Plabeled external 3’-probe or with the internal 5’-probe. Fragment sizes are: CAGGs-GFP: 5’-probe: wt=6.5 kb, targeted=3.8 kb; 3’-probe: wt=6.5 kb, targeted=6.9 kb. D. Phase contrast picture and fluorescence imaging of eGFP in heterozygous or homozygous BG01 clones targeted with an AAVS1 donor plasmid containing a CAGGS driven eGFP cassette and the AAVS1 ZFNs. E. Schematic overview depicting the targeting strategy for the PPP1R12C gene in the AAVS1 locus with a donor construct containing a DOXinducible TetO-eGFP. Probes used for Southern blot analysis are shown as red boxes, the first 3 exons of the PPP1R12C gene in the AAVS1 locus are shown as blue boxes and arrows indicate the genomic site cut by the ZFNs. Donor plasmids used to target the AAVS1 locus are shown above; SA-Puro: splice acceptor sequence followed by a 2A self-cleaving peptide sequence and the puromycin resistance gene, pA: polyadenylation sequence, TetO: Tetracycline response element. F. Phase contrast picture and fluorescence imaging of eGFP in BG01 cells either heterozygous (AAVS1 TetO-GFP+/−) or homozygous (AAVS1-TetOGFP+/+) for the DOX-inducible eGFP cassette targeted to the AAVS1 locus. Cells were transduced with a M2rtTA lentivirus to render the cells DOX responsive. Panel shows colonies before (top) and after FACS assisted subcloning in the presence of DOX (bottom). G. Southern blot analysis of BG01cells targeted with the indicated donor plasmids using the AAVS1 ZFNs. Genomic DNA was digested with SphI and hybridized with the 32P-labeled external 3’-probe or with the internal 5’-probe. H. FACS analysis of AAVS1-TetO-GFP+/− and AAVS1-TetO-GFP+/+ subclones for GFP expression at different concentrations of DOX. BGO1 cells, targeted cells prior to M2rtTA infection, and subcloned GFP responsive cell lines were cultured at different concentrations of DOX and analyzed. All cells were co-stained and analyzed for SSEA4 expression to exclude SSEA4 negative feeder cells from the analysis.
Figure 3. Targeting of PITX3 in hESCs and hiPSCs using ZFNs
A. Schematic overview depicting the targeting strategy for the PITX3 gene. Probes for Southern blot analysis are shown as red boxes, the first exons of the PITX3 locus are shown as blue boxes and arrows indicate the genomic site cut by the ZFN pair#2. Donor plasmids used to target the PITX3 locus are shown above and contained 5’ and 3’ homologous sequences of approximately 800 bp flanking the predicted ZFN pair #2 target site; eGFP: enhanced green fluorescent protein, PGK: human phophoglycerol kinase promotor, PURO: puromycin resistance gene, loxP: loxP sites, pA: polyadenylation sequence. Two constructs that differed only in the orientation of this selection cassette with respect to the PITX3 gene were successfully used to target PITX3 (See also Table 1). B. Southern blot analysis of BG01 cells targeted with the indicated donor plasmid using the PITX3 ZFNs. The right panel shows Southern blot analysis of clones in which the PGK-Puro cassette was removed by transient Cre-recombinase expression. Genomic DNA was digested with HindIII and probed with 32P-labeled external 5’-probe or with the internal 3’-probe. Fragment sizes are: 5’ probe: wt=8.8 kb, targeted=7.4 kb Δ-PGK-Puro=10.5 kb; 3’-probe: wt=8.8 kb, targeted=4.3 kb. C. Southern blot analysis of the hiPSCs targeted with the indicated donor plasmids using the PITX3 ZFNs. Genomic DNA was digested with HindIII and probed with 32P-labeled external 5’ probe or with the internal 3’-probe. Fragment sizes are: 5’ probe: wt=8.8 kb, targeted=7.4 kb; 3’-probe: wt=8.8 kb, targeted=4.3 kb.
Similar articles
- Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells.
Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Zou J, et al. Cell Stem Cell. 2009 Jul 2;5(1):97-110. doi: 10.1016/j.stem.2009.05.023. Epub 2009 Jun 18. Cell Stem Cell. 2009. PMID: 19540188 Free PMC article. - Zinc-finger nuclease enhanced gene targeting in human embryonic stem cells.
Hartley BJ, Fabb SA, Finnin BA, Haynes JM, Pouton CW. Hartley BJ, et al. J Vis Exp. 2014 Aug 23;(90):e51764. doi: 10.3791/51764. J Vis Exp. 2014. PMID: 25177806 Free PMC article. - Genome editing of human embryonic stem cells and induced pluripotent stem cells with zinc finger nucleases for cellular imaging.
Wang Y, Zhang WY, Hu S, Lan F, Lee AS, Huber B, Lisowski L, Liang P, Huang M, de Almeida PE, Won JH, Sun N, Robbins RC, Kay MA, Urnov FD, Wu JC. Wang Y, et al. Circ Res. 2012 Dec 7;111(12):1494-503. doi: 10.1161/CIRCRESAHA.112.274969. Epub 2012 Sep 11. Circ Res. 2012. PMID: 22967807 Free PMC article. - Generation of GFP Reporter Human Induced Pluripotent Stem Cells Using AAVS1 Safe Harbor Transcription Activator-Like Effector Nuclease.
Luo Y, Rao M, Zou J. Luo Y, et al. Curr Protoc Stem Cell Biol. 2014 May 16;29:5A.7.1-18. doi: 10.1002/9780470151808.sc05a07s29. Curr Protoc Stem Cell Biol. 2014. PMID: 24838915 Free PMC article. Review.
Cited by
- Reporter Alleles in hiPSCs: Visual Cues on Development and Disease.
Cotta GC, Teixeira Dos Santos RC, Costa GMJ, Lacerda SMDSN. Cotta GC, et al. Int J Mol Sci. 2024 Oct 13;25(20):11009. doi: 10.3390/ijms252011009. Int J Mol Sci. 2024. PMID: 39456792 Free PMC article. Review. - Biological and biomedical applications of engineered nucleases.
Pan Y, Xiao L, Li AS, Zhang X, Sirois P, Zhang J, Li K. Pan Y, et al. Mol Biotechnol. 2013 Sep;55(1):54-62. doi: 10.1007/s12033-012-9613-9. Mol Biotechnol. 2013. PMID: 23089945 Review. - Chemical and biological approaches to improve the efficiency of homologous recombination in human cells mediated by artificial restriction DNA cutter.
Katada H, Harumoto T, Shigi N, Komiyama M. Katada H, et al. Nucleic Acids Res. 2012 Jun;40(11):e81. doi: 10.1093/nar/gks185. Epub 2012 Feb 23. Nucleic Acids Res. 2012. PMID: 22362741 Free PMC article. - ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering.
Gaj T, Gersbach CA, Barbas CF 3rd. Gaj T, et al. Trends Biotechnol. 2013 Jul;31(7):397-405. doi: 10.1016/j.tibtech.2013.04.004. Epub 2013 May 9. Trends Biotechnol. 2013. PMID: 23664777 Free PMC article. Review. - Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells.
Chang CJ, Bouhassira EE. Chang CJ, et al. Blood. 2012 Nov 8;120(19):3906-14. doi: 10.1182/blood-2012-03-420703. Epub 2012 Sep 21. Blood. 2012. PMID: 23002118 Free PMC article.
References
- Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. - PubMed
- Costa M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. - PubMed
- Irion S, et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-CA087869/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01-HD045022/HD/NICHD NIH HHS/United States
- R37-CA084198/CA/NCI NIH HHS/United States
- R37 CA084198/CA/NCI NIH HHS/United States
- R01 CA087869/CA/NCI NIH HHS/United States
- R01 HD045022/HD/NICHD NIH HHS/United States
- R01 CA084198/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials