DySCo: quantitating associations of membrane proteins using two-color single-molecule tracking - PubMed (original) (raw)
Comment
DySCo: quantitating associations of membrane proteins using two-color single-molecule tracking
Paul D Dunne et al. Biophys J. 2009.
Abstract
We present a general method called dynamic single-molecule colocalization for quantitating the associations of single cell surface molecules labeled with distinct autofluorescent proteins. The chief advantages of the new quantitative approach are that, in addition to stable interactions, it is capable of measuring nonconstitutive associations, such as those induced by the cytoskeleton, and it is applicable to situations where the number of molecules is small.
Figures
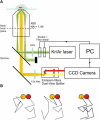
Figure 1
(A) Experimental setup – full details of which can be found in the Supporting Material. T cells expressing proteins labeled with mCherry and Citrine are attached to a glass coverslip coated with nonactivating donkey anti-mouse antibody and the diffusion of the individual molecules on the basal surface of the cell is imaged after exciting simultaneously with overlapped 488 and 568 nm laser beams in total internal reflection geometry. Example raw data can be found as Movie S1. (B) Principle of the DySCo method. Associated molecules track within a short distance of one another for multiple frames (right) whereas unassociated molecules show no correlated motion (left). Unassociated molecules may track together by chance over a short distance (center), but the probability of this occurring for multiple frames is small.
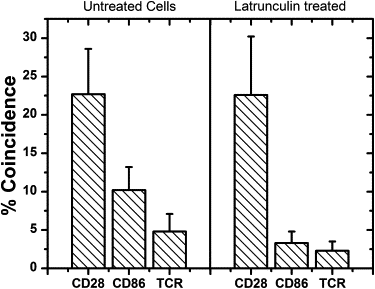
Figure 2
Molecule colocalization for cells with and without latrunculin treatment. See text for details. Data points are mean ± SE.
Comment on
- Fluorescence imaging for monitoring the colocalization of two single molecules in living cells.
Koyama-Honda I, Ritchie K, Fujiwara T, Iino R, Murakoshi H, Kasai RS, Kusumi A. Koyama-Honda I, et al. Biophys J. 2005 Mar;88(3):2126-36. doi: 10.1529/biophysj.104.048967. Epub 2004 Dec 13. Biophys J. 2005. PMID: 15596511 Free PMC article.
Similar articles
- Multidimensional single-molecule imaging in live cells using total-internal-reflection fluorescence microscopy.
Webb SE, Needham SR, Roberts SK, Martin-Fernandez ML. Webb SE, et al. Opt Lett. 2006 Jul 15;31(14):2157-9. doi: 10.1364/ol.31.002157. Opt Lett. 2006. PMID: 16794711 - Fluorescence imaging for monitoring the colocalization of two single molecules in living cells.
Koyama-Honda I, Ritchie K, Fujiwara T, Iino R, Murakoshi H, Kasai RS, Kusumi A. Koyama-Honda I, et al. Biophys J. 2005 Mar;88(3):2126-36. doi: 10.1529/biophysj.104.048967. Epub 2004 Dec 13. Biophys J. 2005. PMID: 15596511 Free PMC article. - Triple-color coincidence analysis: one step further in following higher order molecular complex formation.
Heinze KG, Jahnz M, Schwille P. Heinze KG, et al. Biophys J. 2004 Jan;86(1 Pt 1):506-16. doi: 10.1016/S0006-3495(04)74129-6. Biophys J. 2004. PMID: 14695295 Free PMC article. - Fluorescence fluctuation spectroscopy: a coming of age story.
Van Orden A, Fogarty K, Jung J. Van Orden A, et al. Appl Spectrosc. 2004 May;58(5):122A-137A. doi: 10.1366/000370204774103264. Appl Spectrosc. 2004. PMID: 15165323 Review. No abstract available. - Fluorescent probes for bioimaging applications.
Terai T, Nagano T. Terai T, et al. Curr Opin Chem Biol. 2008 Oct;12(5):515-21. doi: 10.1016/j.cbpa.2008.08.007. Curr Opin Chem Biol. 2008. PMID: 18771748 Review.
Cited by
- Simultaneous Multicolor Single-Molecule Tracking with Single-Laser Excitation via Spectral Imaging.
Huang T, Phelps C, Wang J, Lin LJ, Bittel A, Scott Z, Jacques S, Gibbs SL, Gray JW, Nan X. Huang T, et al. Biophys J. 2018 Jan 23;114(2):301-310. doi: 10.1016/j.bpj.2017.11.013. Biophys J. 2018. PMID: 29401428 Free PMC article. - Raft-based interactions of gangliosides with a GPI-anchored receptor.
Komura N, Suzuki KG, Ando H, Konishi M, Koikeda M, Imamura A, Chadda R, Fujiwara TK, Tsuboi H, Sheng R, Cho W, Furukawa K, Furukawa K, Yamauchi Y, Ishida H, Kusumi A, Kiso M. Komura N, et al. Nat Chem Biol. 2016 Jun;12(6):402-10. doi: 10.1038/nchembio.2059. Epub 2016 Apr 4. Nat Chem Biol. 2016. PMID: 27043189 - Temporal resolution of protein-protein interactions in the live-cell plasma membrane.
Weghuber J, Sunzenauer S, Plochberger B, Brameshuber M, Haselgrübler T, Schütz GJ. Weghuber J, et al. Anal Bioanal Chem. 2010 Aug;397(8):3339-47. doi: 10.1007/s00216-010-3854-x. Epub 2010 Jun 25. Anal Bioanal Chem. 2010. PMID: 20574782 Free PMC article. - Fluorescence techniques to study lipid dynamics.
Sezgin E, Schwille P. Sezgin E, et al. Cold Spring Harb Perspect Biol. 2011 Nov 1;3(11):a009803. doi: 10.1101/cshperspect.a009803. Cold Spring Harb Perspect Biol. 2011. PMID: 21669985 Free PMC article. Review. - Initiation of T cell signaling by CD45 segregation at 'close contacts'.
Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, McColl J, Jönsson P, Palayret M, Harlos K, Coles CH, Jones EY, Lui Y, Huang E, Gilbert RJC, Klenerman D, Aricescu AR, Davis SJ. Chang VT, et al. Nat Immunol. 2016 May;17(5):574-582. doi: 10.1038/ni.3392. Epub 2016 Mar 21. Nat Immunol. 2016. PMID: 26998761 Free PMC article.
References
- Sako Y., Minoghchi S., Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. - PubMed
- Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 2005;6:1253–1262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- 081894/WT_/Wellcome Trust/United Kingdom
- BB/D000467/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources