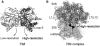Focused functional dynamics of supramolecules by use of a mixed-resolution elastic network model - PubMed (original) (raw)
Focused functional dynamics of supramolecules by use of a mixed-resolution elastic network model
Ozge Kurkcuoglu et al. Biophys J. 2009.
Abstract
The mixed-resolution elastic network model was introduced previously for computing the motions of a structure, which is described at different levels of detail in different parts, for example, with atomistic and residue-level regions. This method has proved to be an efficient tool to explore the collective dynamics of proteins with some atomistic details, which would be difficult to obtain with either conventional full-atom approaches or fully coarse-grained models. Understanding function often requires atomic detail, but not necessarily for the entire structure. In this study, the calculation of the interaction forces between different resolution regions for the hierarchical levels of coarse-graining is further elaborated on in the new approach by considering explicitly the atomic contacts in the crystal structure. The collective dynamics of the enzyme triosephosphate isomerase and its active site together with loop 6 motions are considered in detail. The supramolecular assemblage ribosome and local atomic motions in its "interesting" functional part-the decoding center-are investigated for the low frequency range of the spectrum with high computational efficiency. This new atom-based mixed coarse-graining approach can be effectively used to generate realistic high-resolution conformations of extremely large protein-DNA or RNA complexes by performing energy minimization on structures deformed along the normal modes of the elastic network model. The new model permits focusing on specific functional parts that move in coordination and response to the remainder of the entire structure.
Figures
Figure 1
Cartoon representations of the two model systems used in this study. (A) TIM. (B) Ribosome complexed with A-, P-, E-tRNAs, and mRNA. The dark gray region represented as sticks in TIM, and the black surface of the ribosome complex are considered to be the high-resolution regions in the elastic networks. The active site region on one monomer of TIM and the functional parts of the ribosome complex are thus indicated.
Figure 2
(A) Active sites of the reverse-mapped conformers of TIM with the functional loop 6 are shown in orange (Mod1a), red (Mod1b), cyan (Mod2a), and purple (Mod2b) on monomer A. (B) The anticorrelated twisting motion of the monomers around a rotation axis in the first normal mode. One of the alternative directions of motion is indicated with arrows for some residues. Loop 6 is shown in blue on both monomers of the Mod1a conformer. (C) The closure of loop 6 on monomer B is depicted along the crystal structure (blue), with the reverse-mapped first mode Mod1b (red) from the first ANM and energy minimization cycle, and the reverse-mapped conformation from the second cycle, Mod1b′ (green).
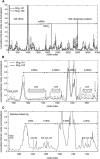
Figure 3
(A) Mean-square fluctuations of the small 30S subunit of the ribosome, averaged over the 10 slowest modes, which match the most collective motions of the complex. Two mixed coarse-grained (mcg) ribosome models with cg levels N/1 (dashed line) and N/5 (solid line) shown using all node centers (atomistic and cg). The individual parts on the subunit are indicated. (B) Mean-square fluctuations for the same ribosome models with the high-resolution (H-R) regions (A- and P-sites and the decoding center A1492, A1493) enlarged to show atomistic detail. (C) Same plot for the uniform cg ribosome where the corresponding cg A- and P-sites and the decoding center are indicated. Note that the node indices are not the same as for the mixed cg models.
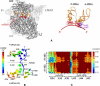
Figure 4
(A) Ratchet-like rotation of the small (30S, in dark gray) and the large (50S, in gray) subunits observed in the third normal mode indicated by arrows for some residues. The high-resolution region is represented in sticks on the mRNA (red), A- and P-tRNAs (orange), and A1492-A1493 (purple). The coarse-graining level for the low-resolution region is N/5. (B) The color-coded mobility in A- and P-sites on the mRNA, tRNAs and nucleotides A1492, A1493, calculated by mean-square fluctuations averaged over the 10 slowest modes. Red color designates high mobility. (C) Correlation plots for the atoms at the decoding center, the codons and anticodons at the A-site.
Similar articles
- Protein dynamics from a NMR perspective: networks of coupled rotators and fractional Brownian dynamics.
Calandrini V, Abergel D, Kneller GR. Calandrini V, et al. J Chem Phys. 2008 Apr 14;128(14):145102. doi: 10.1063/1.2894844. J Chem Phys. 2008. PMID: 18412480 - Loop motions of triosephosphate isomerase observed with elastic networks.
Kurkcuoglu O, Jernigan RL, Doruker P. Kurkcuoglu O, et al. Biochemistry. 2006 Jan 31;45(4):1173-82. doi: 10.1021/bi0518085. Biochemistry. 2006. PMID: 16430213 Free PMC article. - Elastic network models capture the motions apparent within ensembles of RNA structures.
Zimmermann MT, Jernigan RL. Zimmermann MT, et al. RNA. 2014 Jun;20(6):792-804. doi: 10.1261/rna.041269.113. Epub 2014 Apr 23. RNA. 2014. PMID: 24759093 Free PMC article. - NMR spectroscopy on domain dynamics in biomacromolecules.
Shapiro YE. Shapiro YE. Prog Biophys Mol Biol. 2013 Aug;112(3):58-117. doi: 10.1016/j.pbiomolbio.2013.05.001. Epub 2013 May 15. Prog Biophys Mol Biol. 2013. PMID: 23684958 Review. - Controlling proteins through molecular springs.
Zocchi G. Zocchi G. Annu Rev Biophys. 2009;38:75-88. doi: 10.1146/annurev.biophys.050708.133637. Annu Rev Biophys. 2009. PMID: 19416060 Review.
Cited by
- MAVENs: motion analysis and visualization of elastic networks and structural ensembles.
Zimmermann MT, Kloczkowski A, Jernigan RL. Zimmermann MT, et al. BMC Bioinformatics. 2011 Jun 28;12:264. doi: 10.1186/1471-2105-12-264. BMC Bioinformatics. 2011. PMID: 21711533 Free PMC article. - Bridging between NMA and Elastic Network Models: Preserving All-Atom Accuracy in Coarse-Grained Models.
Na H, Jernigan RL, Song G. Na H, et al. PLoS Comput Biol. 2015 Oct 16;11(10):e1004542. doi: 10.1371/journal.pcbi.1004542. eCollection 2015 Oct. PLoS Comput Biol. 2015. PMID: 26473491 Free PMC article. - Global dynamics of proteins: bridging between structure and function.
Bahar I, Lezon TR, Yang LW, Eyal E. Bahar I, et al. Annu Rev Biophys. 2010;39:23-42. doi: 10.1146/annurev.biophys.093008.131258. Annu Rev Biophys. 2010. PMID: 20192781 Free PMC article. Review. - Comparative Analysis of Structural and Dynamical Features of Ribosome Upon Association With mRNA Reveals Potential Role of Ribosomal Proteins.
Bheemireddy S, Sandhya S, Srinivasan N. Bheemireddy S, et al. Front Mol Biosci. 2021 Aug 2;8:654164. doi: 10.3389/fmolb.2021.654164. eCollection 2021. Front Mol Biosci. 2021. PMID: 34409066 Free PMC article. - ClustENM: ENM-Based Sampling of Essential Conformational Space at Full Atomic Resolution.
Kurkcuoglu Z, Bahar I, Doruker P. Kurkcuoglu Z, et al. J Chem Theory Comput. 2016 Sep 13;12(9):4549-62. doi: 10.1021/acs.jctc.6b00319. Epub 2016 Aug 18. J Chem Theory Comput. 2016. PMID: 27494296 Free PMC article.
References
- Levitt M., Sander C., Stern P.S. Protein normal-mode dynamics: trypsin inhibitor, crambin, ribonuclease and lysozyme. J. Mol. Biol. 1985;181:423–447. - PubMed
- McCammon A., Harvey S.C. Cambridge University Press; Cambridge, UK: 1987. Dynamics of Proteins and Nucleic Acids.
- Tirion M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. - PubMed
- Kitao A., Go N. Investigating protein dynamics in collective coordinate space. Curr. Opin. Struct. Biol. 1999;9:164–169. - PubMed
- Berendsen H.J.C., Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000;10:165–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM081680-0/GM/NIGMS NIH HHS/United States
- R01GM072014-5/GM/NIGMS NIH HHS/United States
- R01 GM072014/GM/NIGMS NIH HHS/United States
- R01 GM073095/GM/NIGMS NIH HHS/United States
- R01 GM081680/GM/NIGMS NIH HHS/United States
- R01GM073095-03/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources