Averaged kick maps: less noise, more signal... and probably less bias - PubMed (original) (raw)
Averaged kick maps: less noise, more signal... and probably less bias
Jure Pražnikar et al. Acta Crystallogr D Biol Crystallogr. 2009 Sep.
Abstract
Use of reliable density maps is crucial for rapid and successful crystal structure determination. Here, the averaged kick (AK) map approach is investigated, its application is generalized and it is compared with other map-calculation methods. AK maps are the sum of a series of kick maps, where each kick map is calculated from atomic coordinates modified by random shifts. As such, they are a numerical analogue of maximum-likelihood maps. AK maps can be unweighted or maximum-likelihood (sigma(A)) weighted. Analysis shows that they are comparable and correspond better to the final model than sigma(A) and simulated-annealing maps. The AK maps were challenged by a difficult structure-validation case, in which they were able to clarify the problematic region in the density without the need for model rebuilding. The conclusion is that AK maps can be useful throughout the entire progress of crystal structure determination, offering the possibility of improved map interpretation.
Figures
Figure 1
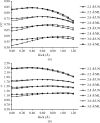
Convergence of AK maps depending on random number seeds. The upper smooth curves show the CCs of pairs of AK maps as a function of the number of maps averaged, whereas the bottom curves plot the CC of each individual map compared with the final map of the series. Three series of AK maps were calculated from three different starting random number seeds to avoid any map repetition. The applied kick size was 0.9 Å at a resolution of 3.0 Å. The crystal structures of P79S stefin B variant and cathepsin H were used as initial models in (a) and (b), respectively.
Figure 2
Map improvement as a function of kick size and model quality. The UN AK and ML AK maps calculated from four molecular models corresponding to four different stages (see Table 2 ▶) in the determination of the crystal structure of cathepsin H. For each model UN AK and ML AK maps were calculated with kick sizes from 0.1 to 1.2 Å from 100 kick maps. Zero kick size corresponds to the UN and ML maps, respectively. In (a) CCs between the final F model map and UN AK and AK maps are shown. (b) shows the average density of the final model atoms in each particular map.
Figure 3
AK maps derived from 100 kick maps were calculated at 3.0 Å resolution for molecular-replacement solutions of cathepsin H (PDB code
8pch
), ammodytin L (
3dih
), stefin B tetramer (
2oct
) and three other structures from the PDB (
2ahn
,
2fy2
and
1twl
), using actinidin, C. atrox phospholipase A2,
1thv
,
1q6x
and
1nde
as search models. The graphs represent the CCs between the F model map of the final refined structures and the AK map (UN AK, triangles; ML AK, circles) of the molecular-replacement solutions at different kick step. The dashed straight line represents the CC between the F model of the final structure and the 2_F_ obs − F calc ML map of the molecular-replacement solution without kicks.
Figure 4
Local map comparison along the chain. UN, ML and AK maps (shown as blue, red and green lines) are compared with the final F model map. CCs were calculated for regions belonging to each individual residue of the final structure and are plotted along the whole chain of ammodytin L (PDB code
3dih
).
Figure 5
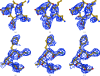
Local map comparison of two regions. (a), (b) and (c) show maps around residues Ile9–Glu13, while (d), (e) and (f) show maps around residues Cys94–Arg98 calculated from a model of ammodytin L (R value 0.37, the same as used to prepare Fig. 4 ▶). The final model is shown in stick representation. The maps in (a) and (d) represent UN maps, those in (b) and (e) represent ML maps and those in (c) and (f) represent AK maps. The maps were generated using data at 3.0 Å resolution and are all shown at a 1.0σ contour level.
Figure 6
AK maps in structure validation. First- and second-generation AK OMIT maps of the region 1–15 are shown on the background of the 4–16 sequence of the
1zen
(a_–_d) and
1b57
(e_–_h) PDB depositions are shown. The first generation of ML AK and UN AK maps are shown in (a) and (e) and in (b) and (f), respectively, and the second-generation ML AL and UN AK maps are shown in (c) and (g) and in (d) and (h), respectively. Kick maps were calculated with a single kick size of 0.8 Å and were averaged 100 times. Maps are contoured at 1.2σ.
Figure 7
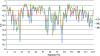
R factors of AK maps plotted against kick size. The plots show the R factors of the maps of cathepsin H generated for Fig. 2 ▶.
Similar articles
- Map-likelihood phasing.
Terwilliger TC. Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2001 Dec;57(Pt 12):1763-75. doi: 10.1107/s0907444901013749. Epub 2001 Nov 21. Acta Crystallogr D Biol Crystallogr. 2001. PMID: 11717488 Free PMC article. - Free kick instead of cross-validation in maximum-likelihood refinement of macromolecular crystal structures.
Pražnikar J, Turk D. Pražnikar J, et al. Acta Crystallogr D Biol Crystallogr. 2014 Dec 1;70(Pt 12):3124-34. doi: 10.1107/S1399004714021336. Epub 2014 Nov 22. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 25478831 Free PMC article. - MAIN software for density averaging, model building, structure refinement and validation.
Turk D. Turk D. Acta Crystallogr D Biol Crystallogr. 2013 Aug;69(Pt 8):1342-57. doi: 10.1107/S0907444913008408. Epub 2013 Jun 13. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23897458 Free PMC article. - Refinement of Atomic Structures Against cryo-EM Maps.
Murshudov GN. Murshudov GN. Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review. - Validating maps from single particle electron cryomicroscopy.
Rosenthal PB, Rubinstein JL. Rosenthal PB, et al. Curr Opin Struct Biol. 2015 Oct;34:135-44. doi: 10.1016/j.sbi.2015.07.002. Epub 2015 Nov 19. Curr Opin Struct Biol. 2015. PMID: 26605834 Review.
Cited by
- RNA Structure Refinement Using the ERRASER-Phenix Pipeline.
Chou FC, Echols N, Terwilliger TC, Das R. Chou FC, et al. Methods Mol Biol. 2016;1320:269-82. doi: 10.1007/978-1-4939-2763-0_17. Methods Mol Biol. 2016. PMID: 26227049 Free PMC article. - Validation and quality assessment of macromolecular structures using complex network analysis.
Pražnikar J, Tomić M, Turk D. Pražnikar J, et al. Sci Rep. 2019 Feb 8;9(1):1678. doi: 10.1038/s41598-019-38658-9. Sci Rep. 2019. PMID: 30737447 Free PMC article. - Conformational flexibility of the ligand-binding domain dimer in kainate receptor gating and desensitization.
Nayeem N, Mayans O, Green T. Nayeem N, et al. J Neurosci. 2011 Feb 23;31(8):2916-24. doi: 10.1523/JNEUROSCI.4771-10.2011. J Neurosci. 2011. PMID: 21414913 Free PMC article. - xMDFF: molecular dynamics flexible fitting of low-resolution X-ray structures.
McGreevy R, Singharoy A, Li Q, Zhang J, Xu D, Perozo E, Schulten K. McGreevy R, et al. Acta Crystallogr D Biol Crystallogr. 2014 Sep;70(Pt 9):2344-55. doi: 10.1107/S1399004714013856. Epub 2014 Aug 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 25195748 Free PMC article. - PHENIX: a comprehensive Python-based system for macromolecular structure solution.
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):213-21. doi: 10.1107/S0907444909052925. Epub 2010 Jan 22. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20124702 Free PMC article.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. - PubMed
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005a). CCP4 Newsl.42, contribution 8.
- Badger, J. (1997). Methods Enzymol.277, 344–352. - PubMed
- Baker, E. N. (1980). J. Mol. Biol.141, 441–484. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials