Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses - PubMed (original) (raw)
Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses
Tony L Goldberg et al. J Virol. 2009 Nov.
Abstract
Nonhuman primates host a plethora of potentially zoonotic microbes, with simian retroviruses receiving heightened attention due to their roles in the origins of human immunodeficiency viruses type 1 (HIV-1) and HIV-2. However, incomplete taxonomic and geographic sampling of potential hosts, especially the African colobines, has left the full range of primate retrovirus diversity unexplored. Blood samples collected from 31 wild-living red colobus monkeys (Procolobus [Piliocolobus] rufomitratus tephrosceles) from Kibale National Park, Uganda, were tested for antibodies to simian immunodeficiency virus (SIV), simian T-cell lymphotrophic virus (STLV), and simian foamy virus (SFV) and for nucleic acids of these same viruses using genus-specific PCRs. Of 31 red colobus tested, 22.6% were seroreactive to SIV, 6.4% were seroreactive to STLV, and 97% were seroreactive to SFV. Phylogenetic analyses of SIV polymerase (pol), STLV tax and long terminal repeat (LTR), and SFV pol and LTR sequences revealed unique SIV and SFV strains and a novel STLV lineage, each divergent from corresponding retroviral lineages previously described in Western red colobus (Procolobus badius badius) or black-and-white colobus (Colobus guereza). Phylogenetic analyses of host mitochondrial DNA sequences revealed that red colobus populations in East and West Africa diverged from one another approximately 4.25 million years ago. These results indicate that geographic subdivisions within the red colobus taxonomic complex exert a strong influence on retroviral phylogeny and that studying retroviral diversity in closely related primate taxa should be particularly informative for understanding host-virus coevolution.
Figures
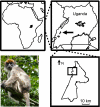
FIG. 1.
Map showing (clockwise from upper left): Uganda within Africa, Kibale National Park within Uganda (arrow), the study site within Kibale National Park (box), and male Kibale red colobus (Procolobus [_Piliocolobus_] rufomitratus tephrosceles).
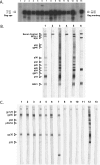
FIG. 2.
Serologic detection of antibodies to simian retroviruses in Kibale red colobus (Procolobus [_Piliocolobus_] rufomitratus tephrosceles). (A) SFV WB analysis. Lanes 2 and 15 are positive control sera from an SFV-infected chimpanzee and African green monkey, respectively; lanes 1 and 14 are negative control sera from an uninfected human; lanes 3 to 13 show the representative reactivities in plasma samples from red colobus monkeys (animals 49, 9, 6, 3, 999, 996, 997, 73, 72, 30, and 29, respectively); The predicted Gag protein sizes for the monkey and ape SFVs are indicated. (B) HTLV WB analysis. Lanes 1 through 3 are from seroreactive and nonreactive plasma samples from red colobus monkeys (animals 49, 72, and 999, respectively); lane 4 is the nonreactive control; lanes 5 and 6 are the HTLV-1- and HTLV-2-reactive positive control antisera, respectively. Reactivity to HTLV-specific proteins is indicated on the left. (C) HIV-2 WB analysis. Lanes 1 to 7 and lanes 8 to 11 show plasma from seroreactive (animals 3, 6, 21, 27, 4, 49, and 67) and nonreactive (animals 11, 31, 54, and 73) red colobus monkeys, respectively; lanes 12 and 13 are assay-positive and -negative control antisera, respectively. Reactivity to HIV-2-specific proteins is indicated on the left.
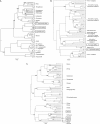
FIG. 3.
Phylogenetic relationships of simian retrovirus sequences from Kibale red colobus (KRC) monkeys using Bayesian inference. Sequences generated in the current study are boxed. 10,000 trees were sampled by an MCMC method under a relaxed clock model, and the maximum clade credibility tree, i.e., the tree with the maximum product of the posterior clade probabilities, was chosen. Scale bars indicate substitutions per site; branch tips are aligned because of the relaxed clock model. Posterior probabilities of >0.70 are provided on each major node. (A) SFV phylogeny of 407-bp polymerase (pol) sequences. Primate species are indicated to the right of the tree. WRC, western red colobus (Procolobus [_Piliocolobus_] badius badius); col, mantled guereza (Colobus guereza). (B) Inferred PTLV-1 LTR phylogeny (511-bp). Corresponding PTLV subtypes or primate hosts are shown to the right of the tree. WRC, western red colobus (Procolobus [_Piliocolobus_] badius badius). (C) SIV pol phylogeny (271 bp). OLC, olive colobus (Procolobus [_Procolobus_] verus); WRC, western red colobus (Procolobus [_Piliocolobus_] badius badius); col, mantled guereza (Colobus guereza); mnd1 and mnd2, mandrill (Mandrillus sphinx); sun, sun-tailed guenon (Cercopithecus solatus); lho, L'Hoest's monkey; smm, sooty mangabey monkey (Cercocebus atys); tan, tantalus guenon (Chlorocebus tantalus); agm, African green monkey (Chlorocebus aethiops); sab, sabaeus guenon (Chlorocebus sabaeus); drl, drill (Mandrillus leucophaeus); rcm, red-capped mangabey (Cercocebus torquatus); cpz, chimpanzee (Pan troglodytes); gsn, greater spot-nosed guenon (Cercopithecus nictitans); asc, red-tailed guenon (Cercopithecus ascanius); mus, mustached guenon (Cercopithecus cephus); mon, mona monkey (Cercopithecus mona); syk, Syke's (or blue) monkey (Cercopithecus mitis); deb, De Brazza's guenon (Cercopithecus neglectus); den, Dent's monkey (Cercopithecus denti); tal, talapoin monkey (Miopithecus talapoin). An asterisk indicates the position of SIV from Temminck's red colobus (Procolobus [_Piliocolobus_] badius temminckii). Sequences of SIVkrc from animals 21 and 40) were excluded from this analysis because they considerably shortened the alignment but clustered with SIVkrc from animal 67 with highly significant bootstrap support (99%) in distance-based trees inferred from the shorter pol alignment (data not shown).
FIG. 4.
Phylogenetic relationships of African colobines using first and second codon positions of NADH3, NADH4L, NADH4, and NADH5 mitochondrial genes (2,548 bp) and Bayesian inference. 10,000 trees were sampled with a MCMC method under a relaxed clock model, and the maximum clade credibility tree was chosen. Branch lengths are proportional to median divergence times in years estimated from the post-burn in trees with the scale at the bottom indicating 5 Ma. Posterior probabilities and median divergence times in Ma and 95% high posterior density intervals (inside brackets) are provided on each major node. Codes used for colobines with SIV infection are shown in parentheses. GenBank accession numbers for sequences included in the analysis are NC002763 (Cebus albifrons), NC001807 (Homo sapiens), NC001807 (Pan troglodytes), NC001992 (Papio hamadryas), EU580083 (Theropithecus gelada), DQ355299 (Presbytis melalophos), EU580051 (Colobus guereza matschiei from Kakamega Forest, Kenya), EU580052 (Colobus guereza occidentalis from Cameroon), EU580082 (Procolobus [_Procolobus_] verus from Tai National Park, Côte d'Ivoire), DQ355301 (Procolobus [_Piliocolobus_] badius badius from Sierra Leone), EU580057 and EU580058 (Procolobus [_Piliocolobus_] badius badius from Tai National Park, Côte d'Ivoire), EU580062 (Procolobus [_Piliocolobus_] kirkii from Zanzibar Island, Tanzania), EU580073 (Procolobus [_Piliocolobus_] pennantii preussi from South Korup National Park, Cameroon), EU580078 (Procolobus [_Piliocolobus_] rufomitratus tephrosceles from Kibale National Park, Uganda), EU580079 (Procolobus [_Piliocolobus_] rufomitratus tephrosceles from Gombe National Park, Tanzania), EU580076 (Procolobus [_Piliocolobus_] temminckii from Abuko Nature Reserve, The Gambia), and EU580077 (Procolobus [_Piliocolobus_] temminckii from Njassang Forest Park, The Gambia).
Similar articles
- High prevalence, coinfection rate, and genetic diversity of retroviruses in wild red colobus monkeys (Piliocolobus badius badius) in Tai National Park, Cote d'Ivoire.
Leendertz SA, Junglen S, Hedemann C, Goffe A, Calvignac S, Boesch C, Leendertz FH. Leendertz SA, et al. J Virol. 2010 Aug;84(15):7427-36. doi: 10.1128/JVI.00697-10. Epub 2010 May 19. J Virol. 2010. PMID: 20484508 Free PMC article. - Dual Simian Foamy Virus/Human Immunodeficiency Virus Type 1 Infections in Persons from Côte d'Ivoire.
Switzer WM, Tang S, Zheng H, Shankar A, Sprinkle PS, Sullivan V, Granade TC, Heneine W. Switzer WM, et al. PLoS One. 2016 Jun 16;11(6):e0157709. doi: 10.1371/journal.pone.0157709. eCollection 2016. PLoS One. 2016. PMID: 27310836 Free PMC article. - Discovery and full genome characterization of two highly divergent simian immunodeficiency viruses infecting black-and-white colobus monkeys (Colobus guereza) in Kibale National Park, Uganda.
Lauck M, Switzer WM, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Taylor B, Shankar A, Ting N, Chapman CA, Friedrich TC, Goldberg TL, O'Connor DH. Lauck M, et al. Retrovirology. 2013 Oct 21;10:107. doi: 10.1186/1742-4690-10-107. Retrovirology. 2013. PMID: 24139306 Free PMC article. - Simian Foamy Virus Co-Infections.
Murray SM, Linial ML. Murray SM, et al. Viruses. 2019 Sep 27;11(10):902. doi: 10.3390/v11100902. Viruses. 2019. PMID: 31569704 Free PMC article. Review. - [Mechanisms of viral emergence and interspecies transmission: the exemple of simian foamy viruses in Central Africa].
Gessain A. Gessain A. Bull Acad Natl Med. 2013 Dec;197(9):1655-67; discussion 1667-8. doi: 10.1016/S0001-4079(19)31387-1. Bull Acad Natl Med. 2013. PMID: 26137812 Free PMC article. Review. French.
Cited by
- High prevalence, coinfection rate, and genetic diversity of retroviruses in wild red colobus monkeys (Piliocolobus badius badius) in Tai National Park, Cote d'Ivoire.
Leendertz SA, Junglen S, Hedemann C, Goffe A, Calvignac S, Boesch C, Leendertz FH. Leendertz SA, et al. J Virol. 2010 Aug;84(15):7427-36. doi: 10.1128/JVI.00697-10. Epub 2010 May 19. J Virol. 2010. PMID: 20484508 Free PMC article. - Discovery and characterization of distinct simian pegiviruses in three wild African Old World monkey species.
Sibley SD, Lauck M, Bailey AL, Hyeroba D, Tumukunde A, Weny G, Chapman CA, O'Connor DH, Goldberg TL, Friedrich TC. Sibley SD, et al. PLoS One. 2014 Jun 11;9(2):e98569. doi: 10.1371/journal.pone.0098569. eCollection 2014. PLoS One. 2014. PMID: 24918769 Free PMC article. - Foamy Viruses, Bet, and APOBEC3 Restriction.
Jaguva Vasudevan AA, Becker D, Luedde T, Gohlke H, Münk C. Jaguva Vasudevan AA, et al. Viruses. 2021 Mar 18;13(3):504. doi: 10.3390/v13030504. Viruses. 2021. PMID: 33803830 Free PMC article. Review. - Contemporary Distribution, Estimated Age, and Prehistoric Migrations of Old World Monkey Retroviruses.
van der Kuyl AC. van der Kuyl AC. Epidemiologia (Basel). 2021 Feb 3;2(1):46-67. doi: 10.3390/epidemiologia2010005. Epidemiologia (Basel). 2021. PMID: 36417189 Free PMC article. Review. - Detection of retroviral super-infection from non-invasive samples.
Goffe AS, Blasse A, Mundry R, Leendertz FH, Calvignac-Spencer S. Goffe AS, et al. PLoS One. 2012;7(5):e36570. doi: 10.1371/journal.pone.0036570. Epub 2012 May 8. PLoS One. 2012. PMID: 22590569 Free PMC article.
References
- Apetrei, C., D. L. Robertson, and P. A. Marx. 2004. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV-infected non-human primates (NHP) in Africa. Front. Biosci. 9:225-254. - PubMed
- Biek, R., A. J. Drummond, and M. Poss. 2006. A virus reveals population structure and recent demographic history of its carnivore host. Science 311:538-541. - PubMed
- Chapman, C. A., and L. J. Chapman. 2002. Foraging challenges of red colobus monkeys: influence of nutrients and secondary compounds. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133:861-875. - PubMed
- Chapman, C. A., and J. E. Lambert. 2000. Habitat alteration and the conservation of African primates: case study of Kibale National Park, Uganda. Am. J. Primatol. 50:169-185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases