Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine - PubMed (original) (raw)
Comparative Study
Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine
Marcus J Claesson et al. PLoS One. 2009.
Abstract
Background: Variations in the composition of the human intestinal microbiota are linked to diverse health conditions. High-throughput molecular technologies have recently elucidated microbial community structure at much higher resolution than was previously possible. Here we compare two such methods, pyrosequencing and a phylogenetic array, and evaluate classifications based on two variable 16S rRNA gene regions.
Methods and findings: Over 1.75 million amplicon sequences were generated from the V4 and V6 regions of 16S rRNA genes in bacterial DNA extracted from four fecal samples of elderly individuals. The phylotype richness, for individual samples, was 1,400-1,800 for V4 reads and 12,500 for V6 reads, and 5,200 unique phylotypes when combining V4 reads from all samples. The RDP-classifier was more efficient for the V4 than for the far less conserved and shorter V6 region, but differences in community structure also affected efficiency. Even when analyzing only 20% of the reads, the majority of the microbial diversity was captured in two samples tested. DNA from the four samples was hybridized against the Human Intestinal Tract (HIT) Chip, a phylogenetic microarray for community profiling. Comparison of clustering of genus counts from pyrosequencing and HITChip data revealed highly similar profiles. Furthermore, correlations of sequence abundance and hybridization signal intensities were very high for lower-order ranks, but lower at family-level, which was probably due to ambiguous taxonomic groupings.
Conclusions: The RDP-classifier consistently assigned most V4 sequences from human intestinal samples down to genus-level with good accuracy and speed. This is the deepest sequencing of single gastrointestinal samples reported to date, but microbial richness levels have still not leveled out. A majority of these diversities can also be captured with five times lower sampling-depth. HITChip hybridizations and resulting community profiles correlate well with pyrosequencing-based compositions, especially for lower-order ranks, indicating high robustness of both approaches. However, incompatible grouping schemes make exact comparison difficult.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
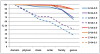
Figure 1. Classification efficiencies at six taxonomic ranks for eight sets of sequences from four samples.
The blue and purple colored dashed lines represent V6 amplicon reads, which have very poor classification efficiencies compared to all V4 amplicon reads, especially at the genus level. The yellow and orange colored dashed lines, representing V4-0.1 amplicon reads, show nearly identical classification efficiencies as the corresponding V4-0.5 amplicon reads.
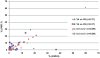
Figure 2. Pearson correlations between genus-classifications for V4 and V6 amplicon sequence datasets, as well as C-V4-0.5 and D-V4-0.5, and C-V4-0.1 and D-V4-0.1 samples.
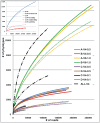
Figure 3. Rarefaction curves at 97% (dotted lines) and 98% levels (solid lines, except for ALL-V4 which has single dots) for all eight datasets including a combination of all V4-0.5/0.1 datasets sequences.
The inset also shows curves for half the A-V6-1.0 reads and the three constituent parts of the C-V4-0.5 reads.
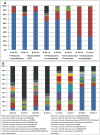
Figure 4. Relative phylum abundance classified with at least 50% bootstrap support (A).
Relative abundance of the 16 most abundant genera classified with at least 50% bootstrap support (B). Genera are labeled according to phylum_class_family_GENUS.
Figure 5. V4 amplicon sequences from the four samples assigned with BLAST and MEGAN.
Pie charts display the relative abundance for each genus. ‘Not assigned’ indicates reads with BLAST hits below the cutoff value.
Figure 6. Comparisons of assignments from the RDP-classifier and MEGAN as ratios of total number of reads for each sample and taxonomic rank.
Blue represents phylum, red class, yellow order, green family, and black genus. Diamonds represent sample A-V4-0.5, squares B-V4-0.5, triangles C-V4-0.5, and circles D-V4-0.5.
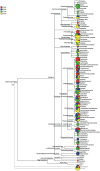
Figure 7. Cluster profiling of HITChip hybridization intensities (left) and number of pyrosequencing reads classified to genus-level with bootstrap support of at least 50% (right).
Figure 8. Comparisons of ratios of HITChip spot intensities and number of pyrosequencing reads for four taxonomic ranks.
Pearson correlations are shown for each rank and sample.
References
- Curtis T. Theory and the microbial world. Environ Microbiol. 2007;9:1. - PubMed
- Woodmansey EJ. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials