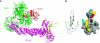Paramyxovirus disruption of interferon signal transduction: STATus report - PubMed (original) (raw)
Review
Paramyxovirus disruption of interferon signal transduction: STATus report
Aparna Ramachandran et al. J Interferon Cytokine Res. 2009 Sep.
Abstract
RNA viruses in the paramyxovirus family have evolved a number of strategies to escape host cell surveillance and antiviral responses. One mechanism exploited by a number of viruses in this family is direct targeting of cytokine-inducible transcription regulators in the STAT family. Diverse members of this large virus family effectively suppress STAT signaling by the actions of their V proteins, or the related proteins derived from alternate viral mRNAs. These viral proteins have distinct means of targeting STATs, resulting in a variety of negative effects on STATs and their signal transduction. Recent developments in understanding STAT targeting will be reviewed.
Figures
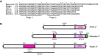
FIG. 1.
Distinct mechanism of STAT inhibition by different paramyxoviruses. (A) Sequence alignment of the highly conserved C-terminal domain (CTD) from various paramyxovirus V proteins mentioned in text. Histidine and 7 cysteine residues and zinc fingers 1 and 2 are indicated. (B) Schematic diagram of PIV5, measles, and Nipah V proteins drawn to scale showing the key features, including the CTD (pink box) and important regions and residues for STAT interaction. (Top) PIV5 V protein. A single region for STAT interaction has not been described, but residue N100 in the N-terminal domain shared between P and V has been shown to provide species-specific interaction with human STAT2. (Middle) Measles V protein. The measles V CTD has a unique extension (green) of unknown significance. The CTD alone is necessary and sufficient for STAT2 interaction. Within the CTD D248 has been found to be important in measles V protein STAT2 interaction. The STAT1 interaction in measles V protein is mediated by residues 110–130 (red box) that includes tyrosine 110 (Y110). (Bottom) Nipah V protein. The Henipavirus CTD is dispensable for STAT interaction. Residues 100–160 (red box) are necessary and sufficient for STAT1 binding, whereas STAT2 association requires STAT1 and a larger region involving residues 100–300. The STAT1-binding domain is also an interaction site for Polo-like kinase 1 (PLK1).
FIG. 2.
Structural representations of paramyxovirus V proteins. (A) The PIV5 V-dependent degradation complex (VDC) complex (PDB accession no. 2B5L) crystal structure (Li and others 2006). The PIV5 V protein (in red) associates with the β-propeller cluster of DDB1 (in green), which is associated with Cullin 4A (in pink) and Roc 1 (in yellow). The crystal structure of PIV5 in complex with DDB1 shows that both the N-terminal domains (NTDs) and C-terminal domains (CTDs) are required to mediate interaction with the components of E3 ligase machinery. The PIV5 V protein is positioned at the substrate-binding cleft of the E3 complex. (B) Structural model of measles V protein CTD showing the surface residues unique to measles V protein that were demonstrated to be important for mediating interaction with STAT2. The measles virus V protein sequence was modeled on the SV5 structure (PDB accession no. 2B5L) by use of the Swiss PDB viewer. Coloration corresponds to the activity level of mutations for each position (Ramachandran and others 2008). Gray represents nonmutated residues, cyan indicates neutral substitutions, yellow represents mutations that retain 30% to 80% activity, and red represents mutations that are >70% defective (F246 and D248) (Ramachandran and others 2008).
Similar articles
- Paramyxovirus activation and inhibition of innate immune responses.
Parks GD, Alexander-Miller MA. Parks GD, et al. J Mol Biol. 2013 Dec 13;425(24):4872-92. doi: 10.1016/j.jmb.2013.09.015. Epub 2013 Sep 20. J Mol Biol. 2013. PMID: 24056173 Free PMC article. Review. - Inhibition of interferon induction and signaling by paramyxoviruses.
Fontana JM, Bankamp B, Rota PA. Fontana JM, et al. Immunol Rev. 2008 Oct;225:46-67. doi: 10.1111/j.1600-065X.2008.00669.x. Immunol Rev. 2008. PMID: 18837775 Review. - Silencing STATs: lessons from paramyxovirus interferon evasion.
Horvath CM. Horvath CM. Cytokine Growth Factor Rev. 2004 Apr-Jun;15(2-3):117-27. doi: 10.1016/j.cytogfr.2004.02.003. Cytokine Growth Factor Rev. 2004. PMID: 15110796 Review. - The regulation of type I interferon production by paramyxoviruses.
Goodbourn S, Randall RE. Goodbourn S, et al. J Interferon Cytokine Res. 2009 Sep;29(9):539-47. doi: 10.1089/jir.2009.0071. J Interferon Cytokine Res. 2009. PMID: 19702509 Free PMC article. Review. - Paramyxovirus strategies for evading the interferon response.
Gotoh B, Komatsu T, Takeuchi K, Yokoo J. Gotoh B, et al. Rev Med Virol. 2002 Nov-Dec;12(6):337-57. doi: 10.1002/rmv.357. Rev Med Virol. 2002. PMID: 12410527 Review.
Cited by
- Viral Respiratory Pathogens and Lung Injury.
Clementi N, Ghosh S, De Santis M, Castelli M, Criscuolo E, Zanoni I, Clementi M, Mancini N. Clementi N, et al. Clin Microbiol Rev. 2021 Mar 31;34(3):e00103-20. doi: 10.1128/CMR.00103-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33789928 Free PMC article. Review. - Reverse genetics of Mononegavirales: How they work, new vaccines, and new cancer therapeutics.
Pfaller CK, Cattaneo R, Schnell MJ. Pfaller CK, et al. Virology. 2015 May;479-480:331-44. doi: 10.1016/j.virol.2015.01.029. Epub 2015 Feb 18. Virology. 2015. PMID: 25702088 Free PMC article. Review. - Virulence of Newcastle disease virus: what is known so far?
Dortmans JC, Koch G, Rottier PJ, Peeters BP. Dortmans JC, et al. Vet Res. 2011 Dec 23;42(1):122. doi: 10.1186/1297-9716-42-122. Vet Res. 2011. PMID: 22195547 Free PMC article. - Recombinant human parainfluenza virus type 2 with mutations in V that permit cellular interferon signaling are not attenuated in non-human primates.
Schaap-Nutt A, D'Angelo C, Amaro-Carambot E, Nolan SM, Davis S, Wise SM, Higgins C, Bradley K, Kim O, Mayor R, Skiadopoulos MH, Collins PL, Murphy BR, Schmidt AC. Schaap-Nutt A, et al. Virology. 2010 Oct 10;406(1):65-79. doi: 10.1016/j.virol.2010.07.011. Epub 2010 Jul 27. Virology. 2010. PMID: 20667570 Free PMC article. - Type I and Type II Interferon Antagonism Strategies Used by Paramyxoviridae: Previous and New Discoveries, in Comparison.
Pisanelli G, Pagnini U, Iovane G, García-Sastre A. Pisanelli G, et al. Viruses. 2022 May 21;14(5):1107. doi: 10.3390/v14051107. Viruses. 2022. PMID: 35632848 Free PMC article. Review.
References
- Alexopoulou L. Holt AC. Medzhitov R. Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Tolllike receptor 3. Nature. 2001;413:732–738. - PubMed
- Caignard G. Bourai M. Jacob Y. Tangy F. Vidalain PO. Inhibition of IFN-alpha/beta signaling by two discrete peptides within measles virus V protein that specifically bind STAT1 and STAT2. Virology. 2009;383:112–120. - PubMed
- Caignard G. Guerbois M. Labernardiere JL. Jacob Y. Jones LM. Wild F. Tangy F. Vidalain PO. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/ beta signaling. Virology. 2007;368:351–362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources