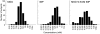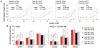Genetic and molecular basis of individual differences in human umami taste perception - PubMed (original) (raw)
Genetic and molecular basis of individual differences in human umami taste perception
Noriatsu Shigemura et al. PLoS One. 2009.
Abstract
Umami taste (corresponds to savory in English) is elicited by L-glutamate, typically as its Na salt (monosodium glutamate: MSG), and is one of five basic taste qualities that plays a key role in intake of amino acids. A particular property of umami is the synergistic potentiation of glutamate by purine nucleotide monophosphates (IMP, GMP). A heterodimer of a G protein coupled receptor, TAS1R1 and TAS1R3, is proposed to function as its receptor. However, little is known about genetic variation of TAS1R1 and TAS1R3 and its potential links with individual differences in umami sensitivity. Here we investigated the association between recognition thresholds for umami substances and genetic variations in human TAS1R1 and TAS1R3, and the functions of TAS1R1/TAS1R3 variants using a heterologous expression system. Our study demonstrated that the TAS1R1-372T creates a more sensitive umami receptor than -372A, while TAS1R3-757C creates a less sensitive one than -757R for MSG and MSG plus IMP, and showed a strong correlation between the recognition thresholds and in vitro dose-response relationships. These results in human studies support the propositions that a TAS1R1/TAS1R3 heterodimer acts as an umami receptor, and that genetic variation in this heterodimer directly affects umami taste sensitivity.
Conflict of interest statement
Competing Interests: Ajinomoto is a product brand name of monosodium glutamate, and the company has interests directly in the sensing of umami. MSG is used in this study and the study could be used to promote the use of MSG and umami.
Figures
Figure 1. Distributions of individual MSG, IMP and MSG+0.5 mM IMP taste recognition thresholds (254 Japanese subjects).
Bin width concentrations correspond to 0.3 log units. The subjects exhibiting recognition threshold over 100 mM MSG, 10 mM IMP, 1.56 mM MSG+0.5 mM IMP are presented as 100<, 10
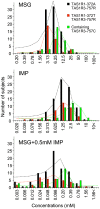
Figure 2. Distributions of individual MSG, IMP and MSG+0.5 mM IMP taste recognition thresholds for three groups defined by TAS1R1-372 and TAS1R3-757 haplotypes.
Black, red and green bars indicate human TAS1R1-372A/TAS1R3-757R homozygotes (n = 84), TAS1R1-372T/TAS1R3-757R homozygotes (n = 37) and homo- and heterozygotes containing the TAS1R3-757C (n = 39), respectively. Black, red and green dotted lines indicate the distribution curves for TAS1R1-372A/TAS1R3-757R homozygotes, TAS1R1-372T/TAS1R3-757R homozygotes and homo- and heterozygotes containing the TAS1R3-757C, respectively, which were obtained by gaussian fit analysis. Bin width concentrations correspond to 0.3 log units. The subjects exhibiting recognition threshold over 100 mM MSG, 10 mM IMP, 1.56 mM MSG+0.5 mM IMP are presented as 100<, 10
Figure 3. Concentration-response relationships of the calcium concentrations in HEK293 cells transfected with human TAS1R1 and TAS1R3 variants after stimulation with increasing MSG, and MSG+0.5 mM IMP.
(A) Dose-response curves of MSG, and MSG+0.5 mM IMP concentration series in cells expressing the TAS1R1/TAS1R3 variants [TAS1R1-372A, -372T, TAS1R3-757R and -757C]. Black and red lines indicate the responses to MSG and MSG+0.5 mM IMP, respectively. Responses have been normalized to those of isoproterenol (ISO, 10 µM), which activate the endogenous βadrenergic receptor . Each point represented the mean (±S.E.) from 10∼18 independent experiments. Half maximal responses (EC50) for TAS1R1/TAS1R3 variants were calculated from the dose-response curves. The X-axis triangles present the EC50 values for MSG (black) and MSG+0.5 mM IMP (red). (B) The responses of TAS1R1/TAS1R3 variants at concentrations of MSG (5, 20, 50 mM) and MSG (0.3, 5, 50 mM)+0.5 mM IMP. The values are means (±S.E.) from 14∼18 independent experiments. Asterisks indicate statistically significant differences (*: p<0.05, †: p<0.01, Fisher's PLSD as a Post-hoc test).
Similar articles
- Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes.
Chen QY, Alarcon S, Tharp A, Ahmed OM, Estrella NL, Greene TA, Rucker J, Breslin PA. Chen QY, et al. Am J Clin Nutr. 2009 Sep;90(3):770S-779S. doi: 10.3945/ajcn.2009.27462N. Epub 2009 Jul 8. Am J Clin Nutr. 2009. PMID: 19587085 Free PMC article. - Variation in umami perception and in candidate genes for the umami receptor in mice and humans.
Shigemura N, Shirosaki S, Ohkuri T, Sanematsu K, Islam AA, Ogiwara Y, Kawai M, Yoshida R, Ninomiya Y. Shigemura N, et al. Am J Clin Nutr. 2009 Sep;90(3):764S-769S. doi: 10.3945/ajcn.2009.27462M. Epub 2009 Jul 22. Am J Clin Nutr. 2009. PMID: 19625681 Free PMC article. Review. - Nonsynonymous single nucleotide polymorphisms in human tas1r1, tas1r3, and mGluR1 and individual taste sensitivity to glutamate.
Raliou M, Wiencis A, Pillias AM, Planchais A, Eloit C, Boucher Y, Trotier D, Montmayeur JP, Faurion A. Raliou M, et al. Am J Clin Nutr. 2009 Sep;90(3):789S-799S. doi: 10.3945/ajcn.2009.27462P. Epub 2009 Jul 1. Am J Clin Nutr. 2009. PMID: 19571223 - Rubemamine and Rubescenamine, Two Naturally Occurring N-Cinnamoyl Phenethylamines with Umami-Taste-Modulating Properties.
Backes M, Obst K, Bojahr J, Thorhauer A, Roudnitzky N, Paetz S, Reichelt KV, Krammer GE, Meyerhof W, Ley JP. Backes M, et al. J Agric Food Chem. 2015 Oct 7;63(39):8694-704. doi: 10.1021/acs.jafc.5b04402. Epub 2015 Sep 24. J Agric Food Chem. 2015. PMID: 26375852 - Molecular insights into human taste perception and umami tastants: A review.
Diepeveen J, Moerdijk-Poortvliet TCW, van der Leij FR. Diepeveen J, et al. J Food Sci. 2022 Apr;87(4):1449-1465. doi: 10.1111/1750-3841.16101. Epub 2022 Mar 17. J Food Sci. 2022. PMID: 35301715 Free PMC article. Review.
Cited by
- Kokumi Taste Active Peptides Modulate Salt and Umami Taste.
Rhyu MR, Song AY, Kim EY, Son HJ, Kim Y, Mummalaneni S, Qian J, Grider JR, Lyall V. Rhyu MR, et al. Nutrients. 2020 Apr 24;12(4):1198. doi: 10.3390/nu12041198. Nutrients. 2020. PMID: 32344605 Free PMC article. - Development of an Enzymatic Biosensor Using Glutamate Oxidase on Organic-Inorganic-Structured, Electrospun Nanofiber-Modified Electrodes for Monosodium Glutamate Detection.
Atilgan H, Unal B, Yalcinkaya EE, Evren G, Atik G, Ozturk Kirbay F, Kilic NM, Odaci D. Atilgan H, et al. Biosensors (Basel). 2023 Mar 28;13(4):430. doi: 10.3390/bios13040430. Biosensors (Basel). 2023. PMID: 37185504 Free PMC article. - A Survey on the Evaluation of Monosodium Glutamate (MSG) Taste in Austria.
Iannilli E, Pötz EL, Hummel T. Iannilli E, et al. Foods. 2024 Dec 25;14(1):22. doi: 10.3390/foods14010022. Foods. 2024. PMID: 39796312 Free PMC article. - Investigating the Effect of Maltodextrins and Degree of Polymerization on Individual Complex Carbohydrate Taste Sensitivity.
Hartley C, Keast RSJ, Bredie WLP. Hartley C, et al. Food Sci Nutr. 2025 Feb 3;13(2):e4751. doi: 10.1002/fsn3.4751. eCollection 2025 Feb. Food Sci Nutr. 2025. PMID: 39906726 Free PMC article. - Intracellular acidification is required for full activation of the sweet taste receptor by miraculin.
Sanematsu K, Kitagawa M, Yoshida R, Nirasawa S, Shigemura N, Ninomiya Y. Sanematsu K, et al. Sci Rep. 2016 Mar 10;6:22807. doi: 10.1038/srep22807. Sci Rep. 2016. PMID: 26960429 Free PMC article.
References
- Yamaguchi S. The synergistic taste effect of monosodium glutamate and disodium 5′-inosinate. J Food Sci. 1967;32:473–478.
- Yoshii K, Yokouchi C, Kurihara K. Synergistic effects of 5′-nucleotides on rat taste responses to various amino acids. Brain Res. 1986;367:45–51. - PubMed
- Lugaz O, Pillias AM, Faurion A. A new specific ageusia: some humans cannot taste l-glutamate. Chem Senses. 2002;27:105–115. - PubMed
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases