Segmentation of center brains and optic lobes in 3D confocal images of adult fruit fly brains - PubMed (original) (raw)
Segmentation of center brains and optic lobes in 3D confocal images of adult fruit fly brains
Shing Chun Benny Lam et al. Methods. 2010 Feb.
Abstract
Automatic alignment (registration) of 3D images of adult fruit fly brains is often influenced by the significant displacement of the relative locations of the two optic lobes (OLs) and the center brain (CB). In one of our ongoing efforts to produce a better image alignment pipeline of adult fruit fly brains, we consider separating CB and OLs and align them independently. This paper reports our automatic method to segregate CB and OLs, in particular under conditions where the signal to noise ratio (SNR) is low, the variation of the image intensity is big, and the relative displacement of OLs and CB is substantial. We design an algorithm to find a minimum-cost 3D surface in a 3D image stack to best separate an OL (of one side, either left or right) from CB. This surface is defined as an aggregation of the respective minimum-cost curves detected in each individual 2D image slice. Each curve is defined by a list of control points that best segregate OL and CB. To obtain the locations of these control points, we derive an energy function that includes an image energy term defined by local pixel intensities and two internal energy terms that constrain the curve's smoothness and length. Gradient descent method is used to optimize this energy function. To improve both the speed and robustness of the method, for each stack, the locations of optimized control points in a slice are taken as the initialization prior for the next slice. We have tested this approach on simulated and real 3D fly brain image stacks and demonstrated that this method can reasonably segregate OLs from CBs despite the aforementioned difficulties.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures
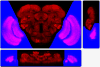
Figure 1
Tri-view of a fruit fly brain and the 3D segmentation surface detected using our deformable model. Painted in blue regions are the segmented optic lobes. The remaining is the center brain. Top-left panel: frontal plane; top-right: sagittal plane; bottom-left: horizontal plane.
Figure 2
(a) Initialization of Chan-Vese level set algorithm on the slice shown in Figure 1. Circles of 30 pixels radii are placed one by one next to each other over the whole image as initialization. Smoothness parameter is 0.1. (b) Segmentation result on the slice shown in Figure 1 using Chan-Vese level set algorithm after 500 iterations followed by morphological hole-filling procedure. The center brain is segmented as several sub-regions.
Figure 2
(a) Initialization of Chan-Vese level set algorithm on the slice shown in Figure 1. Circles of 30 pixels radii are placed one by one next to each other over the whole image as initialization. Smoothness parameter is 0.1. (b) Segmentation result on the slice shown in Figure 1 using Chan-Vese level set algorithm after 500 iterations followed by morphological hole-filling procedure. The center brain is segmented as several sub-regions.
Figure 3
Illustration of deformable model algorithm. The control points ck (red circles) inside the blue box in (a) are magnified and shown in (b) as pointed by the green arrow. For each control point, the respective image energy and internal energy terms are defined (see main text for details)
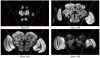
Figure 4
Slice snapshots of a correctly segregated fly brain by (A) deformable model – green line, and (B) shortest path method – red line. Deformable model method correctly segments the OL and CB in all slices while shortest path method gives incorrect result in Slice 68.
Figure 5
Cutting surface in 3D obtained from deformable model method.
Figure 6
Simulation study with different surface shape noise levels. (a) Simulated fly brain in 3D. (b) Slice 71 taken from (a). (c) Zoom in of (b) with different levels of shape surface noise (s) added (from top to bottom: s = 0, s = 0.1, s = 0.3, s = 0.5). Zero-mean Gaussian image noise with variance of 0.1 is added to all images. (d) Segmentation result of brains of different shape noise level using deformable model method with different β values and shortest path method. Diamonds “◆” denote correctly segmented result and crosses “×” denote major segmentations errors found at the corresponding noise level and β value by visual inspection. (e) A major segmentation error is found at the tip of the right OL as indicated by the white arrow.
Figure 7
Examples of minor and major errors. (a) Major error was found on the right side. The green curve cut into a significant region of the right OL. It occurred because several control points were trapped in the local minima in previous slices. (b) Minor error was found on the left side. The green curve cut on a small region belonged to CB as indicated by the red arrow. The error arises because CB is blurry and the gap between OL and CB is very small.
Figure 8
Segmentation of model fruit fly brain of various image noise variances (σ2) and distance betgween OL and CB. Results from deformable model method with different β values and shortest path method are presented. Diamonds “◆” denote correctly segmented results and crosses “×” denote major segmentation errors found from the corresponding OL and CB separation, noise level and β value by visual inspection. Blue masks in the left column represent the segmented regions.
Similar articles
- A principal skeleton algorithm for standardizing confocal images of fruit fly nervous systems.
Qu L, Peng H. Qu L, et al. Bioinformatics. 2010 Apr 15;26(8):1091-7. doi: 10.1093/bioinformatics/btq072. Epub 2010 Feb 19. Bioinformatics. 2010. PMID: 20172944 Free PMC article. - Building generic anatomical models using virtual model cutting and iterative registration.
Xiao M, Soh J, Meruvia-Pastor O, Schmidt E, Hallgrímsson B, Sensen CW. Xiao M, et al. BMC Med Imaging. 2010 Feb 8;10:5. doi: 10.1186/1471-2342-10-5. BMC Med Imaging. 2010. PMID: 20144190 Free PMC article. - A K-means segmentation method for finding 2-D object areas based on 3-D image stacks obtained by confocal microscopy.
Niemistö A, Korpelainen T, Saleem R, Yli-Harja O, Aitchison J, Shmulevich I. Niemistö A, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2007;2007:5559-62. doi: 10.1109/IEMBS.2007.4353606. Annu Int Conf IEEE Eng Med Biol Soc. 2007. PMID: 18003272 - An automatic segmentation algorithm for 3D cell cluster splitting using volumetric confocal images.
Indhumathi C, Cai YY, Guan YQ, Opas M. Indhumathi C, et al. J Microsc. 2011 Jul;243(1):60-76. doi: 10.1111/j.1365-2818.2010.03482.x. Epub 2011 Feb 2. J Microsc. 2011. PMID: 21288236 - Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation.
Rohlfing T, Russakoff DB, Maurer CR Jr. Rohlfing T, et al. IEEE Trans Med Imaging. 2004 Aug;23(8):983-94. doi: 10.1109/TMI.2004.830803. IEEE Trans Med Imaging. 2004. PMID: 15338732
Cited by
- BrainAligner: 3D registration atlases of Drosophila brains.
Peng H, Chung P, Long F, Qu L, Jenett A, Seeds AM, Myers EW, Simpson JH. Peng H, et al. Nat Methods. 2011 Jun;8(6):493-500. doi: 10.1038/nmeth.1602. Epub 2011 May 1. Nat Methods. 2011. PMID: 21532582 Free PMC article.
References
- Brand AH, Perrimon N. Development. 1993;118:401–415. - PubMed
- Lee T, Luo L. Neuron. 1999;22:451–461. - PubMed
- Otsuna H, Ito K. J Comp Neurol. 2006;497:928–958. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases