Cryptic lineages of the genus Escherichia - PubMed (original) (raw)
Cryptic lineages of the genus Escherichia
Seth T Walk et al. Appl Environ Microbiol. 2009 Oct.
Abstract
Extended multilocus sequence typing (MLST) analysis of atypical Escherichia isolates was used to identify five novel phylogenetic clades (CI to CV) among isolates from environmental, human, and animal sources. Analysis of individual housekeeping loci showed that E. coli and its sister clade, CI, remain largely indistinguishable and represent nascent evolutionary lineages. Conversely, clades of similar age (CIII and CIV) were found to be phylogenetically distinct. When all Escherichia lineages (named and unnamed) were evaluated, we found evidence that Escherichia fergusonii has evolved at an accelerated rate compared to E. coli, CI, CIII, CIV, and CV, suggesting that this species is younger than estimated by the molecular clock method. Although the five novel clades were phylogenetically distinct, we were unable to identify a discriminating biochemical marker for all but one of them (CIII) with traditional phenotypic profiling. CIII had a statistically different phenotype from E. coli that resulted from the loss of sucrose and sorbitol fermentation and lysine utilization. The lack of phenotypic distinction has likely hindered the ability to differentiate these clades from typical E. coli, and so their ecological significance and importance for applied and clinical microbiology are yet to be determined. However, our sampling suggests that CIII, CIV, and CV represent environmentally adapted Escherichia lineages that may be more abundant outside the host gastrointestinal tract.
Figures
FIG. 1.
The phylogenetic relationship of novel Escherichia lineages. (A) Position of MLST housekeeping genes relative to the E. coli K-12 MG1655 genome. (B) Split network of isolates showing the phylogenetic position of the five novel clades relative to previously named Escherichia species. (C) UPGMA dendrogram based on the proportion of nucleotide polymorphism differences (_p_-distance).
FIG. 2.
Principal coordinates analysis of all pairwise (strain by strain) chi-square test statistics for Tajima's test of relative evolutionary rates. The analysis ordered isolates into two groups, suggesting that there are two distinct evolutionary rates among the phylogenetic groups of Escherichia species in this study. ECI to ECV, Escherichia clades I to V.
FIG. 3.
Relationship between genetic distance (proportion of nucleotide polymorphisms) and the presence of 27 E. coli pan-genome genes among Escherichia phylogenetic groups. No clear association was found between genetic distance and the percentage of shared loci.
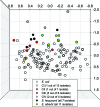
FIG. 4.
NMDS of biochemical profiles for named Escherichia species (green and black circles) and novel Escherichia clades (gray, red, blue, and yellow circles) relative to 692 E. coli isolates (unfilled circles). Only the isolates that represent the 50 most positive and negative positions for each axis are shown. The number of isolates from each clade is given in parentheses. Isolates of the novel clades are highly similar to E. coli (CI to CIV) or completely indistinguishable (CV).
Similar articles
- The "Cryptic" Escherichia.
Walk ST. Walk ST. EcoSal Plus. 2015;6(2):10.1128/ecosalplus.ESP-0002-2015. doi: 10.1128/ecosalplus.ESP-0002-2015. EcoSal Plus. 2015. PMID: 26435255 Free PMC article. Review. - Characterization of the cryptic Escherichia lineages: rapid identification and prevalence.
Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. Clermont O, et al. Environ Microbiol. 2011 Sep;13(9):2468-77. doi: 10.1111/j.1462-2920.2011.02519.x. Epub 2011 Jun 8. Environ Microbiol. 2011. PMID: 21651689 - Characterization of Escherichia fergusonii and Escherichia albertii isolated from water.
Maheux AF, Boudreau DK, Bergeron MG, Rodriguez MJ. Maheux AF, et al. J Appl Microbiol. 2014 Aug;117(2):597-609. doi: 10.1111/jam.12551. Epub 2014 Jun 11. J Appl Microbiol. 2014. PMID: 24849008 - Multiplex polymerase chain reaction for identification of Escherichia coli, Escherichia albertii and Escherichia fergusonii.
Lindsey RL, Garcia-Toledo L, Fasulo D, Gladney LM, Strockbine N. Lindsey RL, et al. J Microbiol Methods. 2017 Sep;140:1-4. doi: 10.1016/j.mimet.2017.06.005. Epub 2017 Jun 6. J Microbiol Methods. 2017. PMID: 28599915 Free PMC article. - Are Escherichia coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing?
Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, Tauschek M. Robins-Browne RM, et al. Front Cell Infect Microbiol. 2016 Nov 18;6:141. doi: 10.3389/fcimb.2016.00141. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27917373 Free PMC article. Review.
Cited by
- Escherichia albertii Pathogenesis.
Gomes TAT, Ooka T, Hernandes RT, Yamamoto D, Hayashi T. Gomes TAT, et al. EcoSal Plus. 2020 Jun;9(1):10.1128/ecosalplus.ESP-0015-2019. doi: 10.1128/ecosalplus.ESP-0015-2019. EcoSal Plus. 2020. PMID: 32588811 Free PMC article. Review. - Molecular diversity of Bacteroidales in fecal and environmental samples and swine-associated subpopulations.
Lamendella R, Li KC, Oerther D, Santo Domingo JW. Lamendella R, et al. Appl Environ Microbiol. 2013 Feb;79(3):816-24. doi: 10.1128/AEM.02535-12. Epub 2012 Nov 16. Appl Environ Microbiol. 2013. PMID: 23160126 Free PMC article. - Household Clustering of Escherichia coli Sequence Type 131 Clinical and Fecal Isolates According to Whole Genome Sequence Analysis.
Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, DebRoy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko EV, Aziz M, Price LB. Johnson JR, et al. Open Forum Infect Dis. 2016 Jun 16;3(3):ofw129. doi: 10.1093/ofid/ofw129. eCollection 2016 Sep. Open Forum Infect Dis. 2016. PMID: 27703993 Free PMC article. - Draft Genome Sequences of Isolates of Diverse Host Origin from the E. coli Reference Center at Penn State University.
Lacher DW, Mammel MK, Gangiredla J, Gebru ST, Barnaba TJ, Majowicz SA, Dudley EG. Lacher DW, et al. Microbiol Resour Announc. 2020 Sep 24;9(39):e01005-20. doi: 10.1128/MRA.01005-20. Microbiol Resour Announc. 2020. PMID: 32972949 Free PMC article. - The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity.
Zhou Z, Alikhan NF, Mohamed K, Fan Y; Agama Study Group; Achtman M. Zhou Z, et al. Genome Res. 2020 Jan;30(1):138-152. doi: 10.1101/gr.251678.119. Epub 2019 Dec 6. Genome Res. 2020. PMID: 31809257 Free PMC article.
References
- Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. Ng, R. Meier, K. Winker, K. K. Ingram, and I. Das. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22:148-155. - PubMed
- Brenner, D. J., B. R. Davis, A. G. Steigerwalt, C. F. Riddle, A. C. McWhorter, S. D. Allen, J. J. Farmer III, Y. Saitoh, and G. R. Fanning. 1982. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov. J. Clin. Microbiol. 15:703-713. - PMC - PubMed
- Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources