Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations - PubMed (original) (raw)
Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations
Yik-Ying Teo et al. Genome Res. 2009 Nov.
Abstract
The Singapore Genome Variation Project (SGVP) provides a publicly available resource of 1.6 million single nucleotide polymorphisms (SNPs) genotyped in 268 individuals from the Chinese, Malay, and Indian population groups in Southeast Asia. This online database catalogs information and summaries on genotype and phased haplotype data, including allele frequencies, assessment of linkage disequilibrium (LD), and recombination rates in a format similar to the International HapMap Project. Here, we introduce this resource and describe the analysis of human genomic variation upon agglomerating data from the HapMap and the Human Genome Diversity Project, providing useful insights into the population structure of the three major population groups in Asia. In addition, this resource also surveyed across the genome for variation in regional patterns of LD between the HapMap and SGVP populations, and for signatures of positive natural selection using two well-established metrics: iHS and XP-EHH. The raw and processed genetic data, together with all population genetic summaries, are publicly available for download and browsing through a web browser modeled with the Generic Genome Browser.
Figures
Figure 1.
Principal component analysis plots of genetic diversity across HapMap, HGDP, and SGVP populations. Each figure represents the genetic diversity seen across the populations considered, with each sample mapped onto a spectrum of genetic variation represented by two axes of variations corresponding to two eigenvectors of the PCA. (A) Individuals from each population in the HapMap and SGVP are represented by a unique color, while samples from HGDP are broadly grouped by geography in which a unique color is assigned to each geographical location. (B) Comparison between CHS and samples from Far East Asia found in the HapMap and HGDP. (C) A plot of the third and fourth axes of variation for the seven populations from HapMap and SGVP. (D) A plot of the first two axes of variation when the PCA is run on only the three Far East Asian populations comprising the Singapore Chinese, HapMap Han Chinese in Beijing, China, and Japanese in Tokyo, Japan. (E) A plot of the first two principal components in a separate analysis within the three SGVP populations. (F) A plot of the second and third principal components within the SGVP populations. The same color scheme has been used in C_–_F; the legend for the color assignment can be found in C.
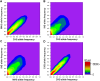
Figure 2.
Allele frequency comparison between pairs of populations. The axes in each figure represent the allele frequencies for each of the two represented populations. For each SNP, we define the minor allele after agglomerating the genotype data from all three SGVP populations and subsequently calculate the frequency of this allele in each population. Twenty allele frequency bins each spanning 0.05 units are constructed for each population, and we tabulate the number of SNPs found in each bin. The intensity of the contour represents the number of SNPs that displayed the corresponding allele frequencies in the two populations, from a low number of SNPs (purple) to a higher number of SNPs (red). The figure panels compare the allelic spectrum among CHS-MAS (A), CHS-INS (B), MAS-INS (C), and CHS-CHB (D).
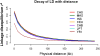
Figure 3.
Decay of LD with distance. Decay of LD as measured by the _r_2 statistic with increasing distance up to 250 kb for each of the HapMap and SGVP populations, where 90 chromosomes were chosen from each population to perform the LD calculation. Only SNPs with minor allele frequencies ≥5% in each population were considered in this analysis.
Figure 4.
LD variation and population-specific recombination rates at CDKAL1. The extent of LD variation between pairs of SGVP and HapMap populations at the CDKAL1 gene, with separate LD heatmaps and recombination rates estimated from genotype data at each population. Population-specific recombination rates are shown except for CHB and JPT, where the same HapMap estimated recombination rates for JPT+CHB are used.
Similar articles
- Natural positive selection and north-south genetic diversity in East Asia.
Suo C, Xu H, Khor CC, Ong RT, Sim X, Chen J, Tay WT, Sim KS, Zeng YX, Zhang X, Liu J, Tai ES, Wong TY, Chia KS, Teo YY. Suo C, et al. Eur J Hum Genet. 2012 Jan;20(1):102-10. doi: 10.1038/ejhg.2011.139. Epub 2011 Jul 27. Eur J Hum Genet. 2012. PMID: 21792231 Free PMC article. - A method for identifying haplotypes carrying the causative allele in positive natural selection and genome-wide association studies.
Ong RT, Liu X, Poh WT, Sim X, Chia KS, Teo YY. Ong RT, et al. Bioinformatics. 2011 Mar 15;27(6):822-8. doi: 10.1093/bioinformatics/btr007. Epub 2011 Jan 6. Bioinformatics. 2011. PMID: 21216773 - SNP identification, linkage disequilibrium, and haplotype analysis for a 200-kb genomic region in a Korean population.
Kim KJ, Lee HJ, Park MH, Cha SH, Kim KS, Kim HT, Kimm K, Oh B, Lee JY. Kim KJ, et al. Genomics. 2006 Nov;88(5):535-40. doi: 10.1016/j.ygeno.2006.03.003. Epub 2006 Aug 17. Genomics. 2006. PMID: 16919420 - Navigating the HapMap.
Barnes MR. Barnes MR. Brief Bioinform. 2006 Sep;7(3):211-24. doi: 10.1093/bib/bbl021. Epub 2006 Jul 28. Brief Bioinform. 2006. PMID: 16877472 Review. - Using haplotype blocks to map human complex trait loci.
Cardon LR, Abecasis GR. Cardon LR, et al. Trends Genet. 2003 Mar;19(3):135-40. doi: 10.1016/S0168-9525(03)00022-2. Trends Genet. 2003. PMID: 12615007 Review.
Cited by
- Pathways-driven sparse regression identifies pathways and genes associated with high-density lipoprotein cholesterol in two Asian cohorts.
Silver M, Chen P, Li R, Cheng CY, Wong TY, Tai ES, Teo YY, Montana G. Silver M, et al. PLoS Genet. 2013 Nov;9(11):e1003939. doi: 10.1371/journal.pgen.1003939. Epub 2013 Nov 21. PLoS Genet. 2013. PMID: 24278029 Free PMC article. - Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry.
Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN. Shriner D, et al. Sci Rep. 2014 Aug 13;4:6055. doi: 10.1038/srep06055. Sci Rep. 2014. PMID: 25116736 Free PMC article. - Mapping the genetic diversity of HLA haplotypes in the Japanese populations.
Saw WY, Liu X, Khor CC, Takeuchi F, Katsuya T, Kimura R, Nabika T, Ohkubo T, Tabara Y, Yamamoto K, Yokota M; Japanese Genome Variation Consortium; Teo YY, Kato N. Saw WY, et al. Sci Rep. 2015 Dec 9;5:17855. doi: 10.1038/srep17855. Sci Rep. 2015. PMID: 26648100 Free PMC article. - Lipidomic profiling of plasma in a healthy Singaporean population to identify ethnic specific differences in lipid levels and associations with disease risk factors.
Begum H, Torta F, Narayanaswamy P, Mundra PA, Ji S, Bendt AK, Saw WY, Teo YY, Soong R, Little PF, Meikle PJ, Wenk MR. Begum H, et al. Clin Mass Spectrom. 2017 Nov 14;6:25-31. doi: 10.1016/j.clinms.2017.11.002. eCollection 2017 Dec. Clin Mass Spectrom. 2017. PMID: 39193417 Free PMC article. - Evidence of recent natural selection on the Southeast Asian deletion (--(SEA)) causing α-thalassemia in South China.
Qiu QW, Wu DD, Yu LH, Yan TZ, Zhang W, Li ZT, Liu YH, Zhang YP, Xu XM. Qiu QW, et al. BMC Evol Biol. 2013 Mar 11;13:63. doi: 10.1186/1471-2148-13-63. BMC Evol Biol. 2013. PMID: 23497175 Free PMC article.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. - PubMed
- Bonnen PE, Pe'er I, Plenge RM, Salit J, Lowe JK, Shapero MH, Lifton RP, Breslow JL, Daly M, Reich DE, et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38:214–217. - PubMed
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Prichard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. - PubMed
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1213. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials