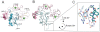Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2 - PubMed (original) (raw)
Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2
Wendy R Gordon et al. PLoS One. 2009.
Abstract
Background: Notch receptors are normally cleaved during maturation by a furin-like protease at an extracellular site termed S1, creating a heterodimer of non-covalently associated subunits. The S1 site lies within a key negative regulatory region (NRR) of the receptor, which contains three highly conserved Lin12/Notch repeats and a heterodimerization domain (HD) that interact to prevent premature signaling in the absence of ligands. Because the role of S1 cleavage in Notch signaling remains unresolved, we investigated the effect of S1 cleavage on the structure, surface trafficking and ligand-mediated activation of human Notch1 and Notch2, as well as on ligand-independent activation of Notch1 by mutations found in human leukemia.
Principal findings: The X-ray structure of the Notch1 NRR after furin cleavage shows little change when compared with that of an engineered Notch1 NRR lacking the S1-cleavage loop. Likewise, NMR studies of the Notch2 HD domain show that the loop containing the S1 site can be removed or cleaved without causing a substantial change in its structure. However, Notch1 and Notch2 receptors engineered to resist S1 cleavage exhibit unexpected differences in surface delivery and signaling competence: S1-resistant Notch1 receptors exhibit decreased, but detectable, surface expression and ligand-mediated receptor activation, whereas S1-resistant Notch2 receptors are fully competent for cell surface delivery and for activation by ligands. Variable dependence on S1 cleavage also extends to T-ALL-associated NRR mutations, as common class 1 mutations display variable decrements in ligand-independent activation when introduced into furin-resistant receptors, whereas a class 2 mutation exhibits increased signaling activity.
Conclusions/significance: S1 cleavage has distinct effects on the surface expression of Notch1 and Notch2, but is not generally required for physiologic or pathophysiologic activation of Notch proteins. These findings are consistent with models for receptor activation in which ligand-binding or T-ALL-associated mutations lead to conformational changes of the NRR that permit metalloprotease cleavage.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
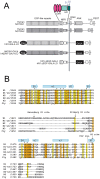
Figure 1. Notch domain organization, constructs used, and sequence alignment.
A. Domain organization and construct design. Sequences derived from Notch1 are white, and sequences from Notch2 are gray. Chimeric Notch1 and Notch2 receptors were created by fusing the extracellular portion of either Notch1 (hN1-Gal4) or Notch2 (hN2/N1-GAL4) with the intracellular portion of Notch1 in which the RAM-ANK domain was replaced with the DNA binding domain of Gal4. Key: FLAG, N-terminal FLAG epitope tag; EGF-like repeats, epidermal growth factor-like repeats; NRR, negative regulatory region consisting of three Lin12/Notch repeats and the heterodimerization domain; TM, transmembrane; ANK, ankyrin repeats; TAD, transcriptional activation domain; PEST, degron domain rich in proline, glutamate, serine, and threonine residues; Gal4, DNA binding domain of the Gal4 transcription factor. B. Multiple sequence alignment of heterodimerization domain sequences of human Notch1–3, Drosophila Notch, and the N1 and N2 deletion constructs (N1-LO and N2-LO, respectively) used in these studies. The amino acid residues are colored according to the clustalW conservation convention: orange- absolute identity in all sequences, yellow- conserved substitution in one or more sequences, grey- semi-conserved substitution in one or more sequences. Alpha helix and beta sheet secondary structural elements of the heterodimerization domain are denoted with light blue cylinders or arrows, respectively. The S2 cleavage site and primary and secondary furin cleavage (S1) sites identified by in vitro furin cleavage of the human Notch1 NRR are marked with arrows. Tumor associated mutations analyzed are denoted by asterisks (point mutations) and an arrow (P12 insertion mutation).
Figure 2. X-ray structure of the Notch1 NRR after furin-cleavage.
A. Ribbon diagram of the structure of the furin-cleaved Notch1 NRR. The LNR modules are in shades of pink. The region of the HD domain that precedes the chain break at the S1 site is colored dark cyan, and the region after the cleavage site is in light blue. Calcium ions are shown as green spheres. The S2 site is indicated by an arrow. B. Overlay of the structure of the S1-cleaved Notch1 NRR (colors as in A) upon that of the Notch1 NRR lacking the S1-cleavage loop (gray; pdb ID code 3eto). C. Close-up view around the S1 site. The cleaved-Notch1 NRR is shown with colored sticks, and the backbone of the loop-deleted Notch1 NRR is shown in white for comparison.
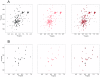
Figure 3. Furin cleavage does not affect the structure of the human Notch2 HD domain.
(A) 15N HSQC spectrum of uniformly 15N-labeled human Notch2 HD domain before (black, left panel) and after (red, center panel) in vitro furin processing. A superposition of the two spectra is also shown (right panel). After acquisition of the initial spectrum of the human Notch2 HD domain (350 mM) at 18°C in 50 mM Bis-Tris (pH 7.0) containing 1 mM CaCl2, 50 mM NaCl, the protein was incubated for 13.5 hrs with 150 µl furin at 37°C. A second spectrum was then taken at 18°C. (B) HSQC spectrum of the human Notch2 HD domain selectively labeled with 15N-Leucine before (black, left panel) and after (red, center panel) furin cleavage. A superposition of the two spectra is also shown (right panel). Spectra in (B) were acquired at pH 6.0; all other conditions were identical to (A).
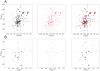
Figure 4. Removal of the S1 loop does not affect the structure of the human Notch2 (N2) HD domain.
(A) Superposition of 15N HSQC spectra of wild-type (black, left panel) and N2-LO (red, center panel) HD domains. A superposition of the two spectra is also shown (right panel). Spectra were acquired at 20°C in 5 mM Bis-Tris (pH 7.0) containing 50 mM NaCl (wild-type) or 5 mM Bis-Tris (pH 6.6) containing 25 mM NaPO4, 50 mM NaCl (N2-LO). (B) Superposition of selectively 15N-Leucine labeled HSQC spectra of wild-type (black, left panel) and N2-LO (red, center panel) HD domains. A superposition of the two spectra is also shown (right panel). Spectra in (B) were acquired at 20°C in 5 mM Bis-Tris (pH 6.8) containing 50 mM NaCl.
Figure 5. Western blots of normal and S1 cleavage-resistant Notch receptors.
(A) Western blot of whole cell extracts of U2OS Flp-in stable cell lines showing tetracycline (Tet)-dependent expression of N1-GAL4 and the loop-deleted N1-GAL4 (hN1-GAL4-LO) chimeric receptors, and (B) Western blot of whole cell extracts of U2OS Flp-in stable cell lines showing tetracycline (Tet)-dependent expression of N2/N1-GAL4 and the loop-deleted N2/N1-GAL4 (N2/N1-GAL4-LO) chimeric receptors. Cells were incubated with Tet (1 mg/ml; +) or carrier alone (−) for 24 hr prior to lysis. Each blot was stained with a rabbit polyclonal antibody specific for the transcriptional activation domain of hN1 . Key: NFL, full-length Notch; NTM, transmembrane subunit of the S1-cleaved receptor. In (B) the lower portion of the blot was stained for α-tubulin as a loading control.
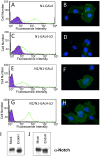
Figure 6. S1-cleavage-resistant Notch receptors are expressed on the cell surface.
Surface receptors were stained with a FITC-conjugated anti-FLAG antibody and detected either by flow cytometry or fluorescence microscopy. (A, B). Surface levels of the N1-Gal4 chimeric receptor, detected by flow cytometry (A) and fluorescence microscopy (B). (C, D). Surface levels of the N1-Gal4-LO chimeric receptor, detected by flow cytometry (C) and fluorescence microscopy (D). (E, F). Surface levels of the N2/N1-Gal4 chimeric receptor, detected by flow cytometry (E) and fluorescence microscopy (F). (G, H). Surface levels of the N2/N1-Gal4-LO chimeric receptor, detected by flow cytometry (G) and fluorescence microscopy (H). In all flow cytometry plots, the green trace corresponds to staining with the FITC-conjugated anti-FLAG antibody, and the black and purple plot shows staining with an isotype-matched control antibody. In the immunofluorescence panels, the surface Notch receptors are stained with anti-FLAG (green), and the nuclei (blue) are stained with DAPI. (I, J) Western blot analysis of Endo H sensitivity. Whole cell lysates from stably transfected U2OS cells expressing the N1-GAL4-LO protein (I) or the N2/N1-GAL4-LO protein (J) were either mock treated (left) or incubated with Endo H glycosidase before analysis (right).
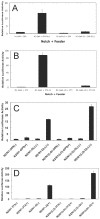
Figure 7. Activation of S1-cleavage resistant Notch receptors by ligands of the Delta and Jagged families.
(A) Isogenic U2OS flp-in cell lines that express N1-Gal4 or N1-Gal4-LO receptors under the control of tetracycline were transfected with a Gal4 responsive luciferase reporter plasmid and co-cultured with OP9 or OP9-Delta-like-1 feeder cells in the presence of tetracycline for 24 hr. Dual luciferase activities were measured in whole cell lysates. In (B), the experiment is identical to that in (A), except that 3T3 or 3T3-Jagged2 feeder cells were used. (C) Isogenic U2OS flp-in cell lines that express N2/N1-Gal4 or N2/N1-Gal4-LO receptors under the control of tetracycline were transfected with a Gal4 responsive luciferase reporter plasmid and co-cultured with OP9 or OP9-Delta-like-1 feeder cells in the presence (+) or absence (−) of tetracycline for 24 hr. Dual luciferase activities were measured in whole cell lysates. In (D), the experiment is identical to that in (C), except that 3T3 or 3T3-Jagged2 feeder cells were used.
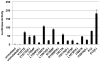
Figure 8. Effect of S1-resistance on ligand-independent activation associated with T-ALL mutations of human Notch1.
U2OS cells were transiently transfected with ΔEGF receptors containing T-ALL associated mutations alone or in cis with the S1-resistant loop-deletion mutation (f-; for furin-resistant). All data points were obtained in triplicate, with error bars indicating the standard deviation of the three replicates.
Similar articles
- Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL.
Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Gordon WR, et al. Blood. 2009 Apr 30;113(18):4381-90. doi: 10.1182/blood-2008-08-174748. Epub 2008 Dec 15. Blood. 2009. PMID: 19075186 Free PMC article. - Human NOTCH2 Is Resistant to Ligand-independent Activation by Metalloprotease Adam17.
Habets RA, Groot AJ, Yahyanejad S, Tiyanont K, Blacklow SC, Vooijs M. Habets RA, et al. J Biol Chem. 2015 Jun 5;290(23):14705-16. doi: 10.1074/jbc.M115.643676. Epub 2015 Apr 27. J Biol Chem. 2015. PMID: 25918160 Free PMC article. - Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes.
Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Malecki MJ, et al. Mol Cell Biol. 2006 Jun;26(12):4642-51. doi: 10.1128/MCB.01655-05. Mol Cell Biol. 2006. PMID: 16738328 Free PMC article. - Notch in Leukemia.
McCarter AC, Wang Q, Chiang M. McCarter AC, et al. Adv Exp Med Biol. 2018;1066:355-394. doi: 10.1007/978-3-319-89512-3_18. Adv Exp Med Biol. 2018. PMID: 30030836 Review. - A brief overview of clinical significance of novel Notch2 regulators.
Pancewicz J. Pancewicz J. Mol Cell Oncol. 2020 Jun 30;7(5):1776084. doi: 10.1080/23723556.2020.1776084. eCollection 2020. Mol Cell Oncol. 2020. PMID: 32944632 Free PMC article. Review.
Cited by
- ADAM17 is critical for multipolar exit and radial migration of neuronal intermediate progenitor cells in mice cerebral cortex.
Li Q, Zhang Z, Li Z, Zhou M, Liu B, Pan L, Ma Z, Zheng Y. Li Q, et al. PLoS One. 2013 Jun 3;8(6):e65703. doi: 10.1371/journal.pone.0065703. Print 2013. PLoS One. 2013. PMID: 23755270 Free PMC article. - Stochastic fluctuations promote ordered pattern formation of cells in the Notch-Delta signaling pathway.
Galbraith M, Bocci F, Onuchic JN. Galbraith M, et al. PLoS Comput Biol. 2022 Jul 21;18(7):e1010306. doi: 10.1371/journal.pcbi.1010306. eCollection 2022 Jul. PLoS Comput Biol. 2022. PMID: 35862460 Free PMC article. - Structural basis for adhesion G protein-coupled receptor Gpr126 function.
Leon K, Cunningham RL, Riback JA, Feldman E, Li J, Sosnick TR, Zhao M, Monk KR, Araç D. Leon K, et al. Nat Commun. 2020 Jan 10;11(1):194. doi: 10.1038/s41467-019-14040-1. Nat Commun. 2020. PMID: 31924782 Free PMC article. - Alagille, Notch, and robustness: why duplicating systems does not ensure redundancy.
Kopan R, Chen S, Liu Z. Kopan R, et al. Pediatr Nephrol. 2014 Apr;29(4):651-7. doi: 10.1007/s00467-013-2661-y. Epub 2013 Nov 24. Pediatr Nephrol. 2014. PMID: 24271660 Free PMC article. Review. - Structural Requirements for Activity of Mind bomb1 in Notch Signaling.
Cao R, Gozlan O, Tveriakhina L, Zhou H, Jiang H, Cole PA, Aster JC, Sprinzak D, Blacklow SC. Cao R, et al. bioRxiv [Preprint]. 2024 Mar 3:2024.03.01.582834. doi: 10.1101/2024.03.01.582834. bioRxiv. 2024. PMID: 38464278 Free PMC article. Updated. Preprint.
References
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. - PubMed
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. - PubMed
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA119070-030003/CA/NCI NIH HHS/United States
- R01 CA092433-05S1/CA/NCI NIH HHS/United States
- P01 CA119070-01A10003/CA/NCI NIH HHS/United States
- R01 CA092433/CA/NCI NIH HHS/United States
- P01 CA119070-039001/CA/NCI NIH HHS/United States
- P01 CA119070-029001/CA/NCI NIH HHS/United States
- P01 CA119070-020003/CA/NCI NIH HHS/United States
- R01 CA092433-05/CA/NCI NIH HHS/United States
- P01 119070/PHS HHS/United States
- P01 CA119070-01A19001/CA/NCI NIH HHS/United States
- R01 CA092433-06A2/CA/NCI NIH HHS/United States
- R01 CA092433-04/CA/NCI NIH HHS/United States
- P01 CA119070/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous