A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis - PubMed (original) (raw)
A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis
Jan Bieschke et al. Protein Sci. 2009 Nov.
Erratum in
- Protein Sci. 2013 Nov;22(11):1688
Abstract
Protein aggregation is a common feature of late onset neurodegenerative disorders, including Alzheimer's disease. In Alzheimer's disease, misassembly of the Abeta peptide is genetically linked to proteotoxicity associated with disease etiology. A reduction in Abeta proteotoxicity is accomplished, in part, by the previously reported Abeta disaggregation and proteolysis activities-under partial control of heat shock factor 1, a transcription factor regulating proteostasis in the cytosol and negatively regulated by insulin growth factor signaling. Herein, we report an improved in vitro assay to quantify recombinant fibrillar Abeta disaggregation kinetics accomplished by the exogenous application of C.elegans extracts. With this assay we demonstrate that the Abeta disaggregation and proteolysis activities of C.elegans are separable. The disaggregation activity found in C.elegans preparations is more heat resistant than the proteolytic activity. Abeta disaggregation in the absence of proteolysis was found to be a reversible process. Future discovery of the molecular basis of the disaggregation and proteolysis activities offers the promise of delaying the age-onset proteotoxicity that leads to neurodegeneration in a spectrum of maladies.
Figures
Figure 1
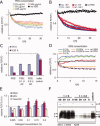
Aβ40 disaggregation activity A: Disaggregation activities of C. elegans PDS as a function of total protein concentration (25–500 μg/mL). Lines show mono-exponential decay function fits to the data (open symbols). B: Disaggregation activities of PDS (20 μg/mL) from Aβ42-expressing (CL for CL2006) and wt (N2) C. elegans fed bacteria expressing hsf-1 RNAi or empty vector controls (EV). Neither the disaggregation rate nor its reduction by hsf-1 downregulation depend on Aβ42 expression. C: PDS-mediated Aβ40 disaggregation is not discernibly inhibited when ATP in the PDS is hydrolyzed by pretreatment with apyrase (AP, 0.1 and 0.5 U, 15 min, 30°C); average relative ThT signals of quintuplicate samples ± SD. D: PDS-mediated (PDS, 25 μg/mL) Aβ40 disaggregation kinetics in the presence of 0.025–0.2% SDS. ThT signals of detergents in buffer were subtracted from the Aβ signals. E: Relative ThT fluorescence signals after 10 h incubation with PDS in the presence of 0.025–0.2% SDS, Tween-20, or NP-40. F: Separation of PDS-mediated disaggregation and proteolysis products of Aβ40 by denaturing PAGE visualized by Western blot analysis (mAb 6E10) of fibrillar Aβ40 before and after PDS treatment. Aβ samples were sonicated for 0, 15, 30, or 60 min before disaggregation. Arrows indicate apparent monomer (M), dimer (D), and trimer (T) bands as well as high-MW aggregate (A) bands. Control: fibrillar Aβ40 incubated for 4 d in the absence of PDS.
Figure 2
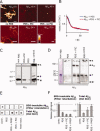
Disaggregation activity is distinct from proteolysis. A: Atomic force microscopy images of fibrillar Aβ42 and Aβ40 incubated in the presence or absence of C. elegans PDS (25 μg/mL) and Roche complete protease inhibitor cocktail (PIC). Scale bars = 200 nm, height scale = 20 nm. Incubation with PDS leads to a loss of readily detectable fibrillar structures both in the presence and absence of PIC. B: Kinetic measurement of Thioflavin T fluorescence of Aβ42 incubated for 75 h with PDS in the presence or absence of PIC. C: Western blot of (mAb 6E10) of monomeric Aβ40 peptide (non agg.), Aβ40 aggregates (agg. control), and the disaggregation products resulting from PDS treatment (25 μg/mL) in the absence or presence of PIC. Arrows indicate apparent monomer (M), dimer (D) and high-MW aggregate (A) bands. D: Coomassie staining reveals monomeric and tetrameric bands of non-aggregated (non agg.) Aβ42. Western blot of Aβ42 aggregates (agg. control), and the disaggregation products resulting from PDS treatment (25 μg/mL) in the absence or presence of PIC. Aβ42 is degraded and proteolyzed after incubation with C. elegans PDS for 72 h in the absence of PIC; bands labeled as in C, in addition a soluble aggregate band was observed (Asol). E: Aβ aliquots from disaggregation assays were analyzed by dot blots and filter retardation assays, respectively, after incubation of Aβ40 for 72 h to quantify the amounts of total Aβ and SDS-resistant Aβ aggregates. F: Quantification of filter retardation assay and dot blot data. Error bars indicate standard deviations in three independent disaggregation experiments. SDS-resistant, aggregated Aβ is reduced after incubation with C. elegans PDS. Total amounts of Aβ peptide remain constant after incubation in the presence of PIC or phenylmethylsulfonylfluoride (PMSF), but decrease in the absence of protease inhibitors.
Figure 3
Disaggregation activity is heat resistant A: Heat-treated C. elegans PDS (25 μg/mL) incubated with fibrillar Aβ40 in the absence of protease inhibitor. PDS was heated to 37, 70, 80 or 95°C for 10 min before incubation with Aβ40 fibrils. B: Dot blot quantification of total Aβ40 using mAB 6E10 after 72 h incubation. C: Aβ40 SDS-resistant aggregate quantification after 72 h incubation (filter retardation, mAB 6E10). Bar graphs show normalized averaged dot blot signals of three independent experiments, error bars indicate standard deviations. Proteolytic activities are inactivated at 70°C, whereas 95°C is needed to inactivate the disaggregation activity D: Fibrillar Aβ42 was incubated with heat-treated C. elegans PDS as in A in the presence or absence of PIC. Disaggregation products were analyzed by Western blotting as in Fig. 2(D). SDS-insoluble aggregate bands (A) and 20 kDa tetramer bands (T) were analyzed quantitatively. Bar graphs show intensities of aggregate (white) and soluble (gray) Aβ42 peptide bands relative to the amounts of Aβ before incubation (control) from four independent experiments, * denotes P < 0.05; ** denotes P < 0.01. E: C. elegans PDS was pretreated without (blue) or with proteinase K (100 μg/mL) (red) for 1 h at 37°C before inactivation of the proteinase K activity by heating to 80°C for 10 min (conditions that leave the disaggregase intact) and then added to fibrillar Aβ40 (15 μ_M_; Aβ monomer).
Figure 4
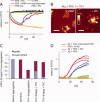
Disaggregation is reversible in the absence of proteolysis. A: C. elegans PDS was incubated with Aβ40 fibrils for 75 h (as in Fig. 1) in the absence (orange) or presence of PIC (blue) or PMSF (red). Inhibition of proteolysis did not prevent disaggregation, but led to reaggregation of Aβ40 upon prolonged incubation. No disaggregation was observed in the presence of heat-inactivated (95°C) PDS in the presence of PIC (green) or in buffer controls (black). B: Atomic force microscopy image of Aβ40 fibrils after 75 h incubation with PDS in the presence of PIC or PMSF, showing re-emergence of fibrils. Scale bars = 200 nm, height scale = 20 nm. C: Aβ40 peptide (15 μ_M_) was separated into soluble and insoluble fractions by centrifugation at 200,000 × g before and after aggregation and after incubation with PDS (25 μg/mL) in the presence or absence of PIC. Both the pellet and the supernatant were treated with HFIP/TFA to denature Aβ40 allowing it to be quantified by HPLC analysis. D: Addition of untreated PDS (5% v/v) to soluble Aβ40 (15 μ_M_) [buffer and incubation as in Fig. 1(A)] prevents aggregation of Aβ into ThT-binding aggregates (triplicate samples, green lines), whereas heat-inactivated PDS (95°C, triplicate samples, red lines), and PDS in the presence of PIC (triplicate samples, blue lines) ultimately allowed Aβ40 aggregation, apparently after the consumption of the disaggregase activity.
Similar articles
- Opposing activities protect against age-onset proteotoxicity.
Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Cohen E, et al. Science. 2006 Sep 15;313(5793):1604-10. doi: 10.1126/science.1124646. Epub 2006 Aug 10. Science. 2006. PMID: 16902091 - HSP110 is a modulator of amyloid beta (Aβ) aggregation and proteotoxicity.
Montresor S, Pigazzini ML, Baskaran S, Sleiman M, Adhikari G, Basilicata L, Secker L, Jacob N, Ehlert Y, Kelkar A, Kalsi GK, Kulkarni N, Spellerberg P, Kirstein J. Montresor S, et al. J Neurochem. 2025 Jan;169(1):e16214. doi: 10.1111/jnc.16214. Epub 2024 Aug 23. J Neurochem. 2025. PMID: 39180255 Free PMC article. - Amyloid-beta (Aβ₁₋₄₂)-induced paralysis in Caenorhabditis elegans is inhibited by the polyphenol quercetin through activation of protein degradation pathways.
Regitz C, Dußling LM, Wenzel U. Regitz C, et al. Mol Nutr Food Res. 2014 Oct;58(10):1931-40. doi: 10.1002/mnfr.201400014. Epub 2014 Jul 28. Mol Nutr Food Res. 2014. PMID: 25066301 - A deuterohemin peptide protects a transgenic Caenorhabditis elegans model of Alzheimer's disease by inhibiting Aβ1-42 aggregation.
Xu J, Yuan Y, Zhang R, Song Y, Sui T, Wang J, Wang C, Chen Y, Guan S, Wang L. Xu J, et al. Bioorg Chem. 2019 Feb;82:332-339. doi: 10.1016/j.bioorg.2018.10.072. Epub 2018 Nov 3. Bioorg Chem. 2019. PMID: 30428413 - Conformations and biological activities of amyloid beta peptide 25-35.
Millucci L, Ghezzi L, Bernardini G, Santucci A. Millucci L, et al. Curr Protein Pept Sci. 2010 Feb;11(1):54-67. doi: 10.2174/138920310790274626. Curr Protein Pept Sci. 2010. PMID: 20201807 Review.
Cited by
- Aging as an event of proteostasis collapse.
Taylor RC, Dillin A. Taylor RC, et al. Cold Spring Harb Perspect Biol. 2011 May 1;3(5):a004440. doi: 10.1101/cshperspect.a004440. Cold Spring Harb Perspect Biol. 2011. PMID: 21441594 Free PMC article. Review. - The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease.
King OD, Gitler AD, Shorter J. King OD, et al. Brain Res. 2012 Jun 26;1462:61-80. doi: 10.1016/j.brainres.2012.01.016. Epub 2012 Jan 21. Brain Res. 2012. PMID: 22445064 Free PMC article. Review. - Aging, protein aggregation, chaperones, and neurodegenerative disorders: mechanisms of coupling and therapeutic opportunities.
Cohen E. Cohen E. Rambam Maimonides Med J. 2012 Oct 31;3(4):e0021. doi: 10.5041/RMMJ.10088. Print 2012 Oct. Rambam Maimonides Med J. 2012. PMID: 23908845 Free PMC article. - Human Hsp70 Disaggregase Reverses Parkinson's-Linked α-Synuclein Amyloid Fibrils.
Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. Gao X, et al. Mol Cell. 2015 Sep 3;59(5):781-93. doi: 10.1016/j.molcel.2015.07.012. Epub 2015 Aug 20. Mol Cell. 2015. PMID: 26300264 Free PMC article. - Polyphenolic glycosides and aglycones utilize opposing pathways to selectively remodel and inactivate toxic oligomers of amyloid β.
Ladiwala AR, Mora-Pale M, Lin JC, Bale SS, Fishman ZS, Dordick JS, Tessier PM. Ladiwala AR, et al. Chembiochem. 2011 Jul 25;12(11):1749-58. doi: 10.1002/cbic.201100123. Epub 2011 Jun 10. Chembiochem. 2011. PMID: 21671331 Free PMC article.
References
- Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. - PubMed
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
- Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat Cell Biol. 2004;6:1054–1061. - PubMed
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. - PubMed
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources