Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements - PubMed (original) (raw)
Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements
Kira S Makarova et al. Biol Direct. 2009.
Abstract
Background: In eukaryotes, RNA interference (RNAi) is a major mechanism of defense against viruses and transposable elements as well of regulating translation of endogenous mRNAs. The RNAi systems recognize the target RNA molecules via small guide RNAs that are completely or partially complementary to a region of the target. Key components of the RNAi systems are proteins of the Argonaute-PIWI family some of which function as slicers, the nucleases that cleave the target RNA that is base-paired to a guide RNA. Numerous prokaryotes possess the CRISPR-associated system (CASS) of defense against phages and plasmids that is, in part, mechanistically analogous but not homologous to eukaryotic RNAi systems. Many prokaryotes also encode homologs of Argonaute-PIWI proteins but their functions remain unknown.
Results: We present a detailed analysis of Argonaute-PIWI protein sequences and the genomic neighborhoods of the respective genes in prokaryotes. Whereas eukaryotic Ago/PIWI proteins always contain PAZ (oligonucleotide binding) and PIWI (active or inactivated nuclease) domains, the prokaryotic Argonaute homologs (pAgos) fall into two major groups in which the PAZ domain is either present or absent. The monophyly of each group is supported by a phylogenetic analysis of the conserved PIWI-domains. Almost all pAgos that lack a PAZ domain appear to be inactivated, and the respective genes are associated with a variety of predicted nucleases in putative operons. An additional, uncharacterized domain that is fused to various nucleases appears to be a unique signature of operons encoding the short (lacking PAZ) pAgo form. By contrast, almost all PAZ-domain containing pAgos are predicted to be active nucleases. Some proteins of this group (e.g., that from Aquifex aeolicus) have been experimentally shown to possess nuclease activity, and are not typically associated with genes for other (putative) nucleases. Given these observations, the apparent extensive horizontal transfer of pAgo genes, and their common, statistically significant over-representation in genomic neighborhoods enriched in genes encoding proteins involved in the defense against phages and/or plasmids, we hypothesize that pAgos are key components of a novel class of defense systems. The PAZ-domain containing pAgos are predicted to directly destroy virus or plasmid nucleic acids via their nuclease activity, whereas the apparently inactivated, PAZ-lacking pAgos could be structural subunits of protein complexes that contain, as active moieties, the putative nucleases that we predict to be co-expressed with these pAgos. All these nucleases are predicted to be DNA endonucleases, so it seems most probable that the putative novel phage/plasmid-defense system targets phage DNA rather than mRNAs. Given that in eukaryotic RNAi systems, the PAZ domain binds a guide RNA and positions it on the complementary region of the target, we further speculate that pAgos function on a similar principle (the guide being either DNA or RNA), and that the uncharacterized domain found in putative operons with the short forms of pAgos is a functional substitute for the PAZ domain.
Conclusion: The hypothesis that pAgos are key components of a novel prokaryotic immune system that employs guide RNA or DNA molecules to degrade nucleic acids of invading mobile elements implies a functional analogy with the prokaryotic CASS and a direct evolutionary connection with eukaryotic RNAi. The predictions of the hypothesis including both the activities of pAgos and those of the associated endonucleases are readily amenable to experimental tests.
Figures
Figure 1
Domain architecture variation in homologs of Argonaute from prokaryotes (pAgos) and eukaryotes (Ago). Structural domains (N-term, L1, PAZ, L2, Mid, PIWI) are projected from the tertiary structure of AaAgo (pdb: 1YVU[35]). Red bars show the inactivated catalytic sites of PIWI domain. Sir2, predicted Sir2 family nuclease domain. APAZ, a domain identified in this work that is associated with pAgos. The domains are shown roughly to scale.
Figure 2
Prokaryotic PIWI-domains: predicted active nucleases and apparently inactivated forms. The multiple sequence alignment includes the core motifs of PIWI domains encompassing the amino acid residues that comprise the (D/E)-(D/E)XK active site. The sequences are denoted by their GI numbers and species names. The positions of the first and the last residues of the aligned region in the corresponding protein are indicated for each sequence. The numbers within the alignment represent poorly conserved inserts that are not shown. The catalytic residues of the D-RD-EXK active site are shown in reverse shading and shown underneath the secondary structure, which corresponds to the solved structure for Pf-Ago (PDB: 1Z25); 'H' indicates α-helix, 'E' indicates extended conformation (β-strand). Sequence identifiers for pAgos that are not associated with other proteins in putative operons are highlighted in bold. The coloring is based on the consensus shown underneath the alignment; 'h' indicates hydrophobic residues (WFYMLIVACTH), 'p' indicates polar residues (EDKRNQHTS), 's' indicates small residues (ACDGNPSTV).
Figure 3
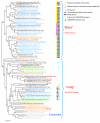
Phylogenetic analysis of PIWI-domains and organization of the predicted pAgo operons. The ML tree is rooted between the (predominantly) PAZ-domain-containing and PAZ-domain- lacking branches. The RELL bootstrap values are indicated (%) for selected major branches. Color code: gray, Eukaryota; orange, Archaea; blue, Proteobacteria, green, Firmicutes; black, other lineages of bacteria. Each organism is denoted by the full systematic name and the Gene Identifier (GI) number. The PDB ID is indicated for those sequences for which tertiary structure is solved. Sequences of short PIWI proteins (that have lost N-terminal part including PAZ domain) but belong to the branch that consists mostly of full size sequences are indicated by "#" symbol. For those PIWI-domain proteins that are associated with genes encoding a nuclease domain, the domain architectures of the pAgo-associated proteins are shown.
Figure 4
Multiple alignment of predicted nuclease domains found in the genomic neighborhoods of pAgo genes. A. Predicted nucleases of the Sir2 family. Numbering of the secondary structure elements corresponds that those reported for PDB: 1j8f[51]. B. (D/E)-(D/E)XK family nucleases. The designations are as in Figure 1. Additional coloring is 'o', hydroxyl-group containing residues (ST); '@', aromatic residues (YWF).
Figure 5
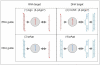
Possible mechanisms of the hypothetical novel prokaryotic systems of defense against mobile elements centered around pAgo compared to the mechanisms of CASS and eukaryotic RNAi. Currently, models (3) and/or (4) are the most likely functional mechanisms for pAgo (see text) but the eukaryotic Ago-like (1) and the prokaryotic CASS-like (2) models cannot be ruled out at this stage. RNA molecules are shown in red and DNA molecules in blue. Circles denote the proteins that form complexes with the guide RNA or DNA. Arrows indicate the directions of the respective processes.
Similar articles
- Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion.
Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V. Zaremba M, et al. Nat Microbiol. 2022 Nov;7(11):1857-1869. doi: 10.1038/s41564-022-01239-0. Epub 2022 Oct 3. Nat Microbiol. 2022. PMID: 36192537 - The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M. Manakova E, et al. Nucleic Acids Res. 2024 Mar 21;52(5):2530-2545. doi: 10.1093/nar/gkad1241. Nucleic Acids Res. 2024. PMID: 38197228 Free PMC article. - A long look at short prokaryotic Argonautes.
Koopal B, Mutte SK, Swarts DC. Koopal B, et al. Trends Cell Biol. 2023 Jul;33(7):605-618. doi: 10.1016/j.tcb.2022.10.005. Epub 2022 Nov 22. Trends Cell Biol. 2023. PMID: 36428175 Review. - Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence.
Koonin EV. Koonin EV. Biol Direct. 2017 Feb 10;12(1):5. doi: 10.1186/s13062-017-0177-2. Biol Direct. 2017. PMID: 28187792 Free PMC article. Review.
Cited by
- A Programmable, DNA-Exclusively-Guided Argonaute DNase and Its Higher Cleavage Specificity Achieved by 5'-Hydroxylated Guide.
Sun S, Xu D, Zhu L, Hu B, Huang Z. Sun S, et al. Biomolecules. 2022 Sep 21;12(10):1340. doi: 10.3390/biom12101340. Biomolecules. 2022. PMID: 36291549 Free PMC article. - Biogenesis and mechanism of action of small non-coding RNAs: insights from the point of view of structural biology.
Costa MC, Leitão AL, Enguita FJ. Costa MC, et al. Int J Mol Sci. 2012;13(8):10268-10295. doi: 10.3390/ijms130810268. Epub 2012 Aug 17. Int J Mol Sci. 2012. PMID: 22949860 Free PMC article. Review. - DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins.
Lisitskaya L, Aravin AA, Kulbachinskiy A. Lisitskaya L, et al. Nat Commun. 2018 Dec 4;9(1):5165. doi: 10.1038/s41467-018-07449-7. Nat Commun. 2018. PMID: 30514832 Free PMC article. Review. - The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes.
Koonin EV, Yutin N. Koonin EV, et al. Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a016188. doi: 10.1101/cshperspect.a016188. Cold Spring Harb Perspect Biol. 2014. PMID: 24691961 Free PMC article. Review. - Argonaute protein as a linker to command center of physiological processes.
Wei K, Wu L, Chen Y, Lin Y, Wang Y, Liu X, Xie D. Wei K, et al. Chin J Cancer Res. 2013 Aug;25(4):430-41. doi: 10.3978/j.issn.1000-9604.2013.08.13. Chin J Cancer Res. 2013. PMID: 23997530 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources