Structure-neurotoxicity relationships of amyloid beta-protein oligomers - PubMed (original) (raw)
Structure-neurotoxicity relationships of amyloid beta-protein oligomers
Kenjiro Ono et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Amyloid beta-protein (Abeta) oligomers may be the proximate neurotoxins in Alzheimer's disease (AD). "Oligomer" is an ill-defined term because many kinds have been reported and they often exist in rapid equilibrium with monomers and higher-order assemblies. We report here results of studies in which specific oligomers have been stabilized structurally, fractionated in pure form, and then studied by using a combination of CD spectroscopy, Thioflavin T fluorescence, EM, atomic force microscopy (AFM), and neurotoxicity assays. Abeta monomers were largely unstructured, but oligomers exhibited order-dependent increases in beta-sheet content. EM and AFM data suggest that dimerization and subsequent monomer addition are processes in which significant and asymmetric monomer conformational changes occur. Oligomer secondary structure and order correlated directly with fibril nucleation activity. Neurotoxic activity increased disproportionately (order dependence >1) with oligomer order. The structure-activity correlations reported here significantly extend our understanding of the conformational dynamics, structure, and relative toxicity of pure Abeta oligomers of specific order.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Stability of purified oligomers. Aβ samples were subjected to PICUP and SDS/PAGE. Individual gel bands were stained with Coomassie blue, excised, and then extracted under alkaline conditions. The extracts then were reconstituted in F12K medium and analyzed by SDS/PAGE and silver staining. Lane 1, cross-linked Aβ immediately after PICUP (cross-linking control). Lane 2, cross-linked Aβ subjected to the entire protocol, but with all bands pooled together (control for unfractionated Aβ subjected to alkaline extraction). Lane 3, monomer band. Lane 4, dimer band. Lane 5, trimer band. Lane 6, tetramer band. Lane 7, a “band-equivalent”-sized piece of gel (“no protein” control). The data are representative of results from each of three independent experiments.
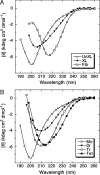
Fig. 2.
Secondary structure dynamics of Aβ assemblies. Uncross-linked (UnXL), cross-linked (XL), or fibrillar (Fib) Aβ (A) or isolated oligomers [monomer (Mo), dimer (Di), trimer (Tr), and tetramer (Te)] (B) were prepared in 10 mM PBS, pH 7.4, and then monitored immediately by CD. Data are representative of three independent experiments.
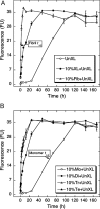
Fig. 3.
Nucleation of Aβ assembly. The nucleation activity of different Aβ preparations was assessed by addition of each preparation to uncross-linked Aβ, which then was incubated for 7 d at 37 °C in 10 mM PBS, pH 7.4. Aliquots were assayed periodically by using ThT. The preparations were uncross-linked Aβ (○, UnXL), 10% (vol/vol) cross-linked Aβ (●, XL), or 10% (vol/vol) sonicated Aβ fibrils (△, Fib) (A) or 10% (vol/vol) Aβ monomer (○, Mo), dimer (●, Di), trimer (△, Tr), or tetramer (▲, Te) (B). Binding is expressed as mean fluorescence [in arbitrary fluorescence units (FU)] ± SE. Data were obtained in three independent experiments. Arrows indicate times at which half maximal ThT binding was observed.
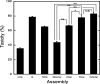
Fig. 4.
LDH activity. Uncross-linked (UnXL), cross-linked (XL), fibrillar, monomeric, and purified oligomeric (dimers, trimers, tetramers) Aβ samples were added at final nominal concentrations of 25 μM to differentiated PC12 cells. LDH activity in the supernatant fluid then was measured after 48 h. Data are representative of that obtained in three independent experiments. Each column represents means ± SE. The statistical significance of the toxicity differences among samples was determined by one-way fractional ANOVA and multiple comparison tests. *, P < 0.01; **, P < 0.001. NS, not significant.
Fig. 5.
Aβ assembly. The data are consistent with an initial oligomerization process in which dimerization involves the self-association of two monomers, each of which exists in a folded (F) state in the dimer. It is not clear when folding from the unfolded (U) state occurs, e.g., before dimerization, contemporaneous with dimerization, or through conformational rearrangement within the dimer. Once the dimer forms, subsequent addition of a monomer to form the trimer involves accommodation (dotted arrow) of the incoming monomer into the dimer structure. The structure of this third monomer within trimers (F*) is different from that of free monomer or of monomers comprising a dimer because the size of the trimer is not thrice that of the monomer or 150% that of the dimer. Each monomer addition past the dimer stage produces the same size increase, and this increase is larger than that observed in dimerization; thus each of these stages would involve monomer accommodation, i.e., F*. It should be noted that every step of peptide oligomerization or fibril formation may not involve simple monomer addition. Other pathways are possible and likely occur (e.g., tetramer formation by dimer association). The scheme presented here illustrates one pathway that is both reasonable and consistent with the experimental data.
Similar articles
- Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer's β-amyloid.
Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Chimon S, et al. Nat Struct Mol Biol. 2007 Dec;14(12):1157-64. doi: 10.1038/nsmb1345. Nat Struct Mol Biol. 2007. PMID: 18059284 - Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity.
Ladiwala AR, Litt J, Kane RS, Aucoin DS, Smith SO, Ranjan S, Davis J, Van Nostrand WE, Tessier PM. Ladiwala AR, et al. J Biol Chem. 2012 Jul 13;287(29):24765-73. doi: 10.1074/jbc.M111.329763. Epub 2012 Apr 30. J Biol Chem. 2012. PMID: 22547072 Free PMC article. - Unmodified and pyroglutamylated amyloid β peptides form hypertoxic hetero-oligomers of unique secondary structure.
Goldblatt G, Cilenti L, Matos JO, Lee B, Ciaffone N, Wang QX, Tetard L, Teter K, Tatulian SA. Goldblatt G, et al. FEBS J. 2017 May;284(9):1355-1369. doi: 10.1111/febs.14058. Epub 2017 Apr 10. FEBS J. 2017. PMID: 28294556 - Amyloid-beta oligomers: their production, toxicity and therapeutic inhibition.
Walsh DM, Klyubin I, Fadeeva JV, Rowan MJ, Selkoe DJ. Walsh DM, et al. Biochem Soc Trans. 2002 Aug;30(4):552-7. doi: 10.1042/bst0300552. Biochem Soc Trans. 2002. PMID: 12196135 Review. - Physicochemical characteristics of soluble oligomeric Abeta and their pathologic role in Alzheimer's disease.
Watson D, Castaño E, Kokjohn TA, Kuo YM, Lyubchenko Y, Pinsky D, Connolly ES Jr, Esh C, Luehrs DC, Stine WB, Rowse LM, Emmerling MR, Roher AE. Watson D, et al. Neurol Res. 2005 Dec;27(8):869-81. doi: 10.1179/016164105X49436. Neurol Res. 2005. PMID: 16354549 Review.
Cited by
- Isotope-edited FTIR reveals distinct aggregation and structural behaviors of unmodified and pyroglutamylated amyloid β peptides.
Goldblatt G, Matos JO, Gornto J, Tatulian SA. Goldblatt G, et al. Phys Chem Chem Phys. 2015 Dec 28;17(48):32149-60. doi: 10.1039/c5cp03343h. Phys Chem Chem Phys. 2015. PMID: 26214017 Free PMC article. - Single-molecule imaging reveals aβ42:aβ40 ratio-dependent oligomer growth on neuronal processes.
Johnson RD, Schauerte JA, Chang CC, Wisser KC, Althaus JC, Carruthers CJ, Sutton MA, Steel DG, Gafni A. Johnson RD, et al. Biophys J. 2013 Feb 19;104(4):894-903. doi: 10.1016/j.bpj.2012.12.051. Biophys J. 2013. PMID: 23442968 Free PMC article. - A genetically immortalized human stem cell line: a promising new tool for Alzheimer's disease therapy.
Puangmalai N, Somani A, Thangnipon W, Ballard C, Broadstock M. Puangmalai N, et al. EXCLI J. 2015 Oct 21;14:1135-14. doi: 10.17179/excli2015-560. eCollection 2015. EXCLI J. 2015. PMID: 27152108 Free PMC article. - Possible Role of Fibrinaloid Microclots in Postural Orthostatic Tachycardia Syndrome (POTS): Focus on Long COVID.
Kell DB, Khan MA, Kane B, Lip GYH, Pretorius E. Kell DB, et al. J Pers Med. 2024 Jan 31;14(2):170. doi: 10.3390/jpm14020170. J Pers Med. 2024. PMID: 38392604 Free PMC article. - Phenolic compounds prevent amyloid β-protein oligomerization and synaptic dysfunction by site-specific binding.
Ono K, Li L, Takamura Y, Yoshiike Y, Zhu L, Han F, Mao X, Ikeda T, Takasaki J, Nishijo H, Takashima A, Teplow DB, Zagorski MG, Yamada M. Ono K, et al. J Biol Chem. 2012 Apr 27;287(18):14631-43. doi: 10.1074/jbc.M111.325456. Epub 2012 Mar 5. J Biol Chem. 2012. PMID: 22393064 Free PMC article.
References
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. - PubMed
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. - PubMed
- Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. - PubMed
- Walsh DM, Selkoe DJ. Aβ oligomers: A decade of discovery. J Neurochem. 2007;101:1172–1184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous