Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex - PubMed (original) (raw)
Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex
Hyuk-Soo Seo et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The Nup84 complex constitutes a key building block in the nuclear pore complex (NPC). Here we present the crystal structure of one of its 7 components, Nup120, which reveals a beta propeller and an alpha-helical domain representing a novel fold. We discovered a previously unidentified interaction of Nup120 with Nup133 and confirmed the physiological relevance in vivo. As mapping of the individual components in the Nup84 complex places Nup120 and Nup133 at opposite ends of the heptamer, our findings indicate a head-to-tail arrangement of elongated Nup84 complexes into a ring structure, consistent with a fence-like coat for the nuclear pore membrane. The attachment site for Nup133 lies at the very end of an extended unstructured region, which allows for flexibility in the diameter of the Nup84 complex ring. These results illuminate important roles of terminal unstructured segments in nucleoporins for the architecture, function, and assembly of the NPC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
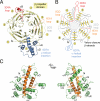
Fig. 1.
Structure of the S. cerevisiae Nup120 NTD. (A) Domain structure. Yellow, β propeller domain; blue, α-helical insertion in the 6D7A loop; green, α-helical domain; gray, α-helical region. The bar above the domain structure denotes the crystallized fragment. (B) Structure of the Nup120 NTD in ribbon representation, color-coded as in (A). A 90°-rotated view is shown on the right.
Fig. 2.
Structural analysis of the Nup120 NTD domains. (A) Ribbon representation of the Nup120 β propeller domain. The 7 blades of the β propeller core (yellow), the location of the disordered 1E1A loop (orange), the 3D4A loop (red), the α-helical insertion in the 6D7A loop (blue), and their secondary structure elements are indicated. (B) Schematic representation of the Nup120 β propeller domain and the locations of its various insertions. (C) Ribbon representation of the Nup120 α-helical domain. The leucine zipper–like core (orange) and the 9 surrounding α-helices (green) are indicated. A 180°-rotated view is shown on the right.
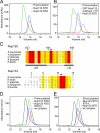
Fig. 3.
Nup120 NTD interacts with the 15 N-terminal residues of Nup133. (A) Gel filtration profiles of Nup120 NTD (blue), Nup133 NTD (red), and the Nup120 NTD·Nup133 NTD complex (green). (B) Gel filtration profiles of Nup120 NTD (blue), the 15 N-terminal residues of Nup133 fused to GST (red), and their complex (green). All proteins were injected at approximately the same concentrations. (C) The invariant Asp-641 of Nup120 and Arg-11 of Nup133 are key residues for complex formation. The location of Asp-641 and Arg-11 are indicated by asterisks in multispecies sequence alignments. (D) Gel filtration profiles of Nup120 NTD D641A mutant (blue) and the Nup133 NTD (red), and the elution profile resulting from incubation of the 2 proteins before injection (green). (E) Gel filtration profiles of Nup120 NTD (blue) and the Nup133 NTD R11A mutant (red), and the elution profile resulting from incubation of the 2 proteins before injection (green). As a reference, the gel filtration profile of the wild-type Nup120 NTD·Nup133 NTD is indicated in black.
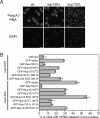
Fig. 4.
Physiological relevance of the Nup120–Nup133 interaction. (A) Detection of poly(A)+ mRNA using an Alexa-647 labeled 50-mer oligo dT FISH probe (Top). Wild-type cells (Left) display a diffuse FISH signal, while _nup120_Δ (Middle) and _nup133_Δ (Right) cells yield strong nuclear signals that coincide with DAPI staining, consistent with poly(A)+ mRNA retention inside the nucleus. (B) Quantitation of nuclear poly(A)+ mRNA retention in _nup120_Δ and _nup133_Δ yeast strains complemented with various Nup120 and Nup133 variants. The percentages refer to the fraction of cells that displayed marked nuclear staining and are derived from 3 independent experiments.
Fig. 5.
Model for the ring formation of the Nup84 complex. (A) Schematic representation of the heptameric complex and the approximate localization of its 7 nups (19). (B) The interaction of Nup120 and Nup133 suggests the intermolecular interaction between 2 heptamers in a head-to-tail fashion mediated by a short stretch at the very N terminus of an extended unstructured region of Nup133. (C) Eight heptamers are arranged in a closed ring with a diameter of ≈1,000 Å in accordance with the NPC dimensions determined by cryo-electron microscopy (8).
Similar articles
- The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture.
Leksa NC, Brohawn SG, Schwartz TU. Leksa NC, et al. Structure. 2009 Aug 12;17(8):1082-91. doi: 10.1016/j.str.2009.06.003. Epub 2009 Jul 2. Structure. 2009. PMID: 19576787 Free PMC article. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Architecture of the symmetric core of the nuclear pore.
Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, Hoelz A. Lin DH, et al. Science. 2016 Apr 15;352(6283):aaf1015. doi: 10.1126/science.aaf1015. Epub 2016 Apr 14. Science. 2016. PMID: 27081075 Free PMC article. - Integrative structure and functional anatomy of a nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, Chemmama IE, Pellarin R, Echeverria I, Shivaraju M, Chaudhury AS, Wang J, Williams R, Unruh JR, Greenberg CH, Jacobs EY, Yu Z, de la Cruz MJ, Mironska R, Stokes DL, Aitchison JD, Jarrold MF, Gerton JL, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP. Kim SJ, et al. Nature. 2018 Mar 22;555(7697):475-482. doi: 10.1038/nature26003. Epub 2018 Mar 14. Nature. 2018. PMID: 29539637 Free PMC article. - 3D ultrastructure of the nuclear pore complex.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Curr Opin Cell Biol. 2012 Feb;24(1):86-91. doi: 10.1016/j.ceb.2011.12.011. Epub 2012 Jan 11. Curr Opin Cell Biol. 2012. PMID: 22244612 Free PMC article. Review. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
References
- Hoelz A, Blobel G. Cell biology: Popping out of the nucleus. Nature. 2004;432:815–816. - PubMed
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. - PubMed
- Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. - PubMed
- Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials