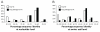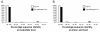Functional characterization of the antibiotic resistance reservoir in the human microflora - PubMed (original) (raw)
Functional characterization of the antibiotic resistance reservoir in the human microflora
Morten O A Sommer et al. Science. 2009.
Abstract
To understand the process by which antibiotic resistance genes are acquired by human pathogens, we functionally characterized the resistance reservoir in the microbial flora of healthy individuals. Most of the resistance genes we identified using culture-independent sampling have not been previously identified and are evolutionarily distant from known resistance genes. By contrast, nearly half of the resistance genes we identified in cultured aerobic gut isolates (a small subset of the gut microbiome) are identical to resistance genes harbored by major pathogens. The immense diversity of resistance genes in the human microbiome could contribute to future emergence of antibiotic resistance in human pathogens.
Figures
Figure 1
Distributions of (A) nucleotide identities and (B) amino acid identities for 93 resistance genes identified from DNA extracted directly from saliva and fecal samples to the most similar resistance gene from any organism (white bars) as well as the most similar resistance gene harbored by a pathogenic isolate (black bars) in GenBank (Table S3)(24). Two resistance genes with no significant similarity to sequences in GenBank are not shown.
Figure 2
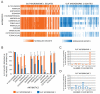
Antibiotic resistance profiles of cultured aerobic gut microbiome isolates. (A) Heat map displaying resistance profiles of 572 aerobic bacterial isolates obtained across 3 different sampling times from two human gut microbiomes. 7436 growth measurements of aerobic cultured microbiome isolates after 24 hrs at 37 °C in Luria Broth containing one of 13 antibiotics at concentrations between 20 and 100 mg/mL that prevent the growth of wild type E.coli (Table S1) are displayed as linear color-scaled bars. White denotes no growth, and color intensity is proportional to growth in the presence of antibiotic, scaled to growth in the absence of antibiotic per individual isolate. (B) Percentage of aerobic gut isolates resistant to each of 13 antibiotics. Each data point represents the mean number of isolates resistant to each antibiotic, and error bars represent the standard deviation of this mean value from each of the three sampling times. Histogram depicting the distribution of the number of different antibiotics that aerobic (C) gut microbiome 1 and (D) gut microbiome 2 isolates are resistant to.
Figure 3
Distributions of (A) nucleotide identities and (B) amino acid identities for 114 resistance genes identified from cultured aerobic gut isolates to the most similar resistance gene from any organism (white bars) as well as the most similar resistance gene harbored by a pathogenic isolate (black bars) in GenBank (Table S4)(24). One resistance gene with no significant similarity to sequences in GenBank is not shown.
Figure 4
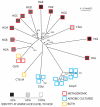
The phylogenetic relationship of unique beta-lactamases derived from gut and oral microbiomes from healthy humans is displayed as an unrooted neighbor-joining tree (24). Except for nodes indicated, bootstrap values = 1000. Scale bar is in fixed amino acid substitutions per sequence position. Border colors of squares denote sequences derived from cultured aerobic gut isolates (blue), metagenomic DNA (red) or both (yellow). Internal shading of each square represents percentage amino acid identity to the most similar sequence in GenBank, with a linear gradient between 100% identity (white) and 35% identity (black). Sequence groups are labeled according to standard nomenclature (Table 1)(24, 32).
Similar articles
- The human microbiome harbors a diverse reservoir of antibiotic resistance genes.
Sommer MO, Church GM, Dantas G. Sommer MO, et al. Virulence. 2010 Jul-Aug;1(4):299-303. doi: 10.4161/viru.1.4.12010. Virulence. 2010. PMID: 21178459 - Antibiotic resistomes of healthy pig faecal metagenomes.
Joyce A, McCarthy CGP, Murphy S, Walsh F. Joyce A, et al. Microb Genom. 2019 May;5(5):e000272. doi: 10.1099/mgen.0.000272. Epub 2019 May 15. Microb Genom. 2019. PMID: 31091181 Free PMC article. - Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner.
Montassier E, Valdés-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E. Montassier E, et al. Nat Microbiol. 2021 Aug;6(8):1043-1054. doi: 10.1038/s41564-021-00920-0. Epub 2021 Jul 5. Nat Microbiol. 2021. PMID: 34226711 Free PMC article. - Horizontal transfer of antibiotic resistance genes in the human gut microbiome.
McInnes RS, McCallum GE, Lamberte LE, van Schaik W. McInnes RS, et al. Curr Opin Microbiol. 2020 Feb;53:35-43. doi: 10.1016/j.mib.2020.02.002. Epub 2020 Mar 3. Curr Opin Microbiol. 2020. PMID: 32143027 Review. - What's new in antibiotic resistance? Focus on beta-lactamases.
Babic M, Hujer AM, Bonomo RA. Babic M, et al. Drug Resist Updat. 2006 Jun;9(3):142-56. doi: 10.1016/j.drup.2006.05.005. Epub 2006 Aug 8. Drug Resist Updat. 2006. PMID: 16899402 Review.
Cited by
- Genomic insights into virulence, antimicrobial resistance, and adaptation acumen of Escherichia coli isolated from an urban environment.
Saini P, Bandsode V, Singh A, Mendem SK, Semmler T, Alam M, Ahmed N. Saini P, et al. mBio. 2024 Mar 13;15(3):e0354523. doi: 10.1128/mbio.03545-23. Epub 2024 Feb 20. mBio. 2024. PMID: 38376265 Free PMC article. - Crowdsourced Data Indicate Widespread Multidrug Resistance in Skin Flora of Healthy Young Adults.
Freeman S, Okoroafor NO, Gast CM, Koval M, Nowowiejski D, O'Connor E, Harrington RD, Parks JW, Fang FC. Freeman S, et al. J Microbiol Biol Educ. 2016 Mar 1;17(1):172-82. doi: 10.1128/jmbe.v17i1.1008. eCollection 2016 Mar. J Microbiol Biol Educ. 2016. PMID: 27047615 Free PMC article. - Antimicrobial resistance characteristics and fitness of Gram-negative fecal bacteria from volunteers treated with minocycline or amoxicillin.
Kirchner M, Mafura M, Hunt T, Abu-Oun M, Nunez-Garcia J, Hu Y, Weile J, Coates A, Card R, Anjum MF. Kirchner M, et al. Front Microbiol. 2014 Dec 17;5:722. doi: 10.3389/fmicb.2014.00722. eCollection 2014. Front Microbiol. 2014. PMID: 25566232 Free PMC article. - Resistance Genes and Genetic Elements Associated with Antibiotic Resistance in Clinical and Commensal Isolates of Streptococcus salivarius.
Chaffanel F, Charron-Bourgoin F, Libante V, Leblond-Bourget N, Payot S. Chaffanel F, et al. Appl Environ Microbiol. 2015 Jun 15;81(12):4155-63. doi: 10.1128/AEM.00415-15. Epub 2015 Apr 10. Appl Environ Microbiol. 2015. PMID: 25862227 Free PMC article. - Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials.
Martínez JL. Martínez JL. Front Microbiol. 2012 Jan 13;3:1. doi: 10.3389/fmicb.2012.00001. eCollection 2012. Front Microbiol. 2012. PMID: 22275914 Free PMC article. No abstract available.
References
- Walsh C. Nature. 2000 Aug;406:775. - PubMed
- Alekshun MN, Levy SB. Cell. 2007 Mar;128:1037. - PubMed
- CDC . Centers for Disease Control and Prevention - Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus, 2006. 2006.
- CDC Centers for Disease Control and Prevention HIV/AIDS Surveillance Report, 2006. 2008;18
- Ochman H, Lawrence JG, Groisman EA. Nature. 2000 May;405:299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials