Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi - PubMed (original) (raw)
Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi
Nelson C Lau et al. EMBO J. 2009.
Erratum in
- EMBO J. 2009 Nov 4;28(21):3458
Abstract
Piwi proteins and Piwi-interacting RNAs (piRNAs) are essential for germ cell development, but analysis of the molecular mechanisms of these ribonucleoproteins remains challenging in most animal germ cells. To address this challenge, we systematically characterized Xiwi, a Xenopus Piwi homologue, and piRNAs from Xenopus eggs and oocytes. We used the large size of Xenopus eggs to analyze small RNAs at the single cell level, and find abundant piRNAs and large piRNA clusters in the Xenopus tropicalis genome, some of which resemble the Drosophila piRNA-generating flamenco locus. Although most piRNA clusters are expressed simultaneously in an egg, individual frogs show distinct profiles of cluster expression. Xiwi is associated with microtubules and the meiotic spindle, and is localized to the germ plasm--a cytoplasmic determinant of germ cell formation. Xiwi associates with translational regulators in an RNA-dependent manner, but Xenopus tudor interacts with Xiwi independently of RNA. Our study adds insight to piRNA transcription regulation by showing that individual animals can have differential piRNA expression profiles. We suggest that in addition to regulating transposable elements, Xiwi may function in specifying RNA localization in vertebrate oocytes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
Xiwi expression during Xenopus oogenesis. (A) Quantitative western blot of Xiwi in staged X. tropicalis oocytes and X. tropicalis and X. laevis eggs. Each lane contains the indicated number of oocytes or eggs. Expression levels were calculated from standard curves of the recombinant Xiwi fragment used to generate the antibody. Our antibody shows lower affinity for Xiwi from X. laevis (data not shown), which explains the apparent lower levels of Xiwi in X. laevis egg extract. (B) Quantification of Xiwi expression levels from quantitative western blots carried out in triplicate. (C) Xiwi was detected in X. tropicalis testes extracts or X. laevis eggs that were manually dissected into animal and vegetal halves. Xiwi concentration in testes extracts was comparable with that observed in stage I oocytes, and approximately equal amounts of Xiwi were observed in the animal and vegetal halves of a X. laevis egg. (D) Stage VI X. laevis oocytes were manually dissected to separate the germinal vesicle from the cytoplasm and the resulting fractions were probed for Xiwi, Xtr, ePAB (as a control for cytoplasm), and histone H4 (as a control for nuclear protein).
Figure 2
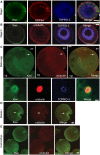
Xiwi localizes to germ plasm and microtubules throughout Xenopus oogenesis. (A, B) Xiwi was detected by immunofluorescence in X. laevis stage I (A) or stage III (B) eggs, and was compared with the localization of α-tubulin or GRP-94, a marker for the Balbiani body. (C) Xiwi was detected by immunofluorescence in mature meiosis II-arrested X. laevis eggs and observed to localize to the granules in the vegetal (Vg) pole of the egg and to co-localize with α-tubulin at the meiosis II spindle in the animal pole (An) of the egg. Higher magnification view of Xiwi localization to the meiotic spindle is shown. (D) Stage VI X. laevis eggs were matured in vitro using progesterone and Xiwi localization was examined during meiosis I. (E) X. tropicalis 8-cell stage embryos were stained for Xiwi and α-tubulin. Xiwi was found to localize predominately to germ plasm islands. TOPRO-3 was used to stain nucleic acids. Scale bars are 100 μm in all panels except (C) lower panel, which is 10 μm. Arrows point specifically to localization of Xiwi and α-tubulin in the meiotic spindle.
Figure 3
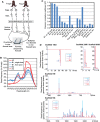
Xenopus piRNA genomics. (A) Size distribution of small RNA reads from Xiwi IP libraries sequenced on the Illumina GA-II (left), and microtubule and egg extract libraries from Xenopus laevis and Xenopus tropicalis sequenced on the 454 Life Science GA-FLX (right). (B) Statistics and annotations of three select small RNA libraries. The complete list of statistics and annotations are detailed in Supplementary Table S1. (C) X. tropicalis piRNA partial cluster maps from the three scaffolds with the greatest number of uniquely mapping reads. Normalized reads are superimposed, and positive and negative reads refer to plus strand and minus strand matches, respectively. Gap, gene and TE annotations were selected from the UCSC genome browser for X. tropicalis genome build 4.1, using the Xenrep5 database to plot the TE annotations (Hinrichs et al, 2006).
Figure 4
Single cell small RNA deep sequencing reveals differential piRNA cluster expression (A). Experimental scheme for multiplex sequencing of small RNA libraries from single X. tropicalis eggs. (B) Size distribution of reads from single-egg small RNA libraries. (C) Pairwise correlation comparison of genome-wide small RNA profiles in which the larger library is normalized to the size of the smaller library. Small RNA profiles measured the polarity and normalized number of uniquely mapping reads in 2 kb windows (see Materials and methods). (D) Partial clusters with similar or different expression between single frog eggs. The cluster on scaffold 1403 has similar normalized expression levels between eggs of frog A (marked with arrows) and frog B. Clusters on scaffold 2356 and 5393 are more highly expressed in eggs from frog B, whereas clusters on scaffold 199 and 166 are more highly expressed in frog A.
Figure 5
Xiwi associates with proteins involved in mRNA translation. (A) Xiwi was immunoprecipitated (IPed) from X. laevis egg extracts and separated by SDS–PAGE. Associated proteins were excised from the gel and identified by mass spectrometry. Protein identities of the top hits are indicated next to the excised band. (B) X. laevis egg extract was cross-linked by the addition of formaldehyde. Xiwi was then IPed, and associated proteins were identified by mass spectrometry. The top hits are indicated next to each band. (C) Xiwi-associated proteins were IPed from X. laevis egg extracts and blotted for the presence of Xiwi. For the ePAB experiment, recombinant GFP–ePAB was added to extracts and immunoprecipitated using anti-GFP antibodies. Anti-Xiwi antibodies cross react with both GFP–ePAB and GFP because the Xiwi antigen containing sequence present in the pET30 expression vector that is present in GFP–ePAB and GFP. (D) RNA was purified from Xiwi IPs and separated on an agarose gel. Xiwi IPs were compared with control IgG IPs. Size markers indicated are DNA markers. (E) Xiwi was IPed from X. laevis egg extracts and the IPs were washed with or without RNaseA to digest associated RNAs, then blotted for associated proteins.
Figure 6
Xiwi co-localizes with Xtr in the germ plasm. Xiwi localization was examined during X. laevis oogenesis and compared with the localization of the xPiwi-associated protein Xtr. (A) In stage I oocytes, Xiwi was observed to localize to the Balbiani body at which it co-localized with Xtr. (B) In stage III oocytes Xiwi localization coalesced into a thin ribbon at the vegetal cortex of the oocytes in which it also co-localized with Xtr. In stage VI and mature eggs, Xiwi localized to granular structures present in the vegetal pole of the egg, reminiscent of the germ plasm. At no stage during oogenesis did we observe Xiwi localization to the nucleus as judged by a lack of co-staining with histone H4. Scale bars are 100 μm in A and B and 10 μm in (C).
Similar articles
- Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome.
Toombs JA, Sytnikova YA, Chirn GW, Ang I, Lau NC, Blower MD. Toombs JA, et al. RNA. 2017 Apr;23(4):504-520. doi: 10.1261/rna.058859.116. Epub 2016 Dec 28. RNA. 2017. PMID: 28031481 Free PMC article. - Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline.
Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Wilczynska A, et al. RNA. 2009 Feb;15(2):337-45. doi: 10.1261/rna.1422509. RNA. 2009. PMID: 19144913 Free PMC article. - Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm.
Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. Vourekas A, et al. Nature. 2016 Mar 17;531(7594):390-394. doi: 10.1038/nature17150. Epub 2016 Mar 7. Nature. 2016. PMID: 26950602 Free PMC article. - piRNA clusters as a main source of small RNAs in the animal germline.
Olovnikov IA, Kalmykova AI. Olovnikov IA, et al. Biochemistry (Mosc). 2013 Jun;78(6):572-84. doi: 10.1134/S0006297913060035. Biochemistry (Mosc). 2013. PMID: 23980884 Review. - A novel epigenetic mechanism in Drosophila somatic cells mediated by Piwi and piRNAs.
Lin H, Yin H. Lin H, et al. Cold Spring Harb Symp Quant Biol. 2008;73:273-81. doi: 10.1101/sqb.2008.73.056. Epub 2009 Mar 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19270080 Free PMC article. Review.
Cited by
- Small Non-Coding RNAs as New Biomarkers to Evaluate the Quality of the Embryo in the IVF Process.
Toporcerová S, Špaková I, Šoltys K, Klepcová Z, Kľoc M, Bohošová J, Trachtová K, Peterová L, Mičková H, Urdzík P, Mareková M, Slabý O, Rabajdová M. Toporcerová S, et al. Biomolecules. 2022 Nov 14;12(11):1687. doi: 10.3390/biom12111687. Biomolecules. 2022. PMID: 36421701 Free PMC article. - The Biogenesis and Functions of piRNAs in Human Diseases.
Wu X, Pan Y, Fang Y, Zhang J, Xie M, Yang F, Yu T, Ma P, Li W, Shu Y. Wu X, et al. Mol Ther Nucleic Acids. 2020 Sep 4;21:108-120. doi: 10.1016/j.omtn.2020.05.023. Epub 2020 May 23. Mol Ther Nucleic Acids. 2020. PMID: 32516734 Free PMC article. Review. - A kinesin Klp10A mediates cell cycle-dependent shuttling of Piwi between nucleus and nuage.
Venkei ZG, Choi CP, Feng S, Chen C, Jacobsen SE, Kim JK, Yamashita YM. Venkei ZG, et al. PLoS Genet. 2020 Mar 13;16(3):e1008648. doi: 10.1371/journal.pgen.1008648. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32168327 Free PMC article. - Identification and differential expression of piRNAs in the gonads of Amur sturgeon (Acipenser schrenckii).
Yuan L, Li L, Zhang X, Jiang H, Chen J. Yuan L, et al. PeerJ. 2019 Apr 12;7:e6709. doi: 10.7717/peerj.6709. eCollection 2019. PeerJ. 2019. PMID: 31106045 Free PMC article. - Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Yao Q, Cao G, Li M, Wu B, Zhang X, Zhang T, Guo J, Yin H, Shi L, Chen J, Yu X, Zheng L, Ma J, Su YQ. Yao Q, et al. Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):11250-11255. doi: 10.1073/pnas.1809744115. Epub 2018 Oct 17. Proc Natl Acad Sci U S A. 2018. PMID: 30333187 Free PMC article.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 - PubMed
- Aravin AA, Hannon GJ, Brennecke J (2007a) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007b) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 HD057298/HD/NICHD NIH HHS/United States
- R00 HD057298/HD/NICHD NIH HHS/United States
- R01 GM086434/GM/NIGMS NIH HHS/United States
- R01 GM086434-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases