LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans - PubMed (original) (raw)
. 2009 Oct;16(10):1016-20.
doi: 10.1038/nsmb.1675. Epub 2009 Aug 27.
Affiliations
- PMID: 19713957
- PMCID: PMC2988485
- DOI: 10.1038/nsmb.1675
LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans
Nicolas J Lehrbach et al. Nat Struct Mol Biol. 2009 Oct.
Abstract
The let-7 microRNA (miRNA) is an ultraconserved regulator of stem cell differentiation and developmental timing and a candidate tumor suppressor. Here we show that LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 processing in Caenorhabditis elegans. We demonstrate that lin-28 is necessary and sufficient to block let-7 activity in vivo; LIN-28 directly binds let-7 pre-miRNA to prevent Dicer processing. Moreover, we have identified a poly(U) polymerase, PUP-2, which regulates the stability of LIN-28-blockaded let-7 pre-miRNA and contributes to LIN-28-dependent regulation of let-7 during development. We show that PUP-2 and LIN-28 interact directly, and that LIN-28 stimulates uridylation of let-7 pre-miRNA by PUP-2 in vitro. Our results demonstrate that LIN-28 and let-7 form an ancient regulatory switch, conserved from nematodes to humans, and provide insight into the mechanism of LIN-28 action in vivo. Uridylation by a PUP-2 ortholog might regulate let-7 and additional miRNAs in other species. Given the roles of Lin28 and let-7 in stem cell and cancer biology, we propose that such poly(U) polymerases are potential therapeutic targets.
Figures
Figure 1. A quantitative assay reveals post-transcriptional regulation of the let-7 miRNA by lin-28
(a,b) Schematic of the pharynx based assay of let-7 activity. (c) Fluorescence images of animals carrying the let-7 sensor transgene at L1 larval and adult stages. Both GFP and mCherry are strongly expressed. d Fluorescence images of animals carrying both the let-7 sensor and myo-2::let-7 transgenes at L1 larval and adult stages. GFP is specifically and robustly downregulated in adults, but not in L1 larvae. Scale bar shows 20 μm. e Fluorescence images showing that lin-28 mutants downregulate let-7 sensor GFP at the L1 stage in a myo-2::let-7 dependent fashion. This effect is not reversed in a lin-46 mutant background. Scale bar shows 20 μm. f Fluorescence images showing that a myo-2::lin-28::unc-54 transgene is sufficient to block let-7 activity in adults carrying let-7 sensor and myo-2::let-7 transgenes. Scale bar shows 20 μm.
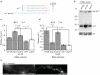
Figure 2. pup-2 regulates let-7 processing in a _lin-28_-dependent fashion
a let-7 is uridylated in vivo. Frequency of unmodified and modified let-7* molecules identified by high-throughput sequencing. b Representative northern blot showing _pup-2_-dependent regulation of pre-let-7. 5 _μ_g of total RNA from control, lin-28(RNAi), and pup-2(RNAi) myo-2::let-7 L2 larvae was loaded. U6 was used as a loading control. c,d Quantification of relative pre-let-7 (c), and let-7 (d) abundance in lin-28(RNAi), pup-2(RNAi) and cid-1(RNAi) myo-2::let-7 L2 and L4 larvae from northern blotting experiments. Mean fold change relative to empty vector control samples is shown. _P_-values from Students’ _t_-tests indicated; n = 4. Error bars show standard error of the mean. e Fluorescence image showing the seam cell defect observed in pup-2(RNAi) adults. A DLG-1-mCherry fusion marks seam cell boundaries. Upper panel; wild-type with continuous seam. Lower panel; pup-2(RNAi) with incompletely fused seam. Arrows indicate sites of failed fusion. Scale bar shows 20 μm.
Figure 3. LIN-28 interacts with PUP-2 and promotes uridylation of pre-let-7 by PUP-2
a Co-Immunoprecipitation of PUP-2 and LIN-28 expressed in HEK293T cells. b GST pull-down assay demonstrating a direct interaction of GST-LIN-28 and PUP-2 in vitro. c In vitro uridylation assay showing that PUP-2 uridylates pre-let-7 in a LIN-28 dependent fashion.
Similar articles
- Regulation of pre-miRNA processing.
Lehrbach NJ, Miska EA. Lehrbach NJ, et al. Adv Exp Med Biol. 2010;700:67-75. Adv Exp Med Biol. 2010. PMID: 21627031 Review. - A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans.
Stefani G, Chen X, Zhao H, Slack FJ. Stefani G, et al. RNA. 2015 May;21(5):985-96. doi: 10.1261/rna.045542.114. Epub 2015 Mar 24. RNA. 2015. PMID: 25805859 Free PMC article. - LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans.
Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. Van Wynsberghe PM, et al. Nat Struct Mol Biol. 2011 Mar;18(3):302-8. doi: 10.1038/nsmb.1986. Epub 2011 Feb 6. Nat Struct Mol Biol. 2011. PMID: 21297634 Free PMC article. - lin-28 controls the succession of cell fate choices via two distinct activities.
Vadla B, Kemper K, Alaimo J, Heine C, Moss EG. Vadla B, et al. PLoS Genet. 2012;8(3):e1002588. doi: 10.1371/journal.pgen.1002588. Epub 2012 Mar 22. PLoS Genet. 2012. PMID: 22457637 Free PMC article. - An elegant miRror: microRNAs in stem cells, developmental timing and cancer.
Nimmo RA, Slack FJ. Nimmo RA, et al. Chromosoma. 2009 Aug;118(4):405-18. doi: 10.1007/s00412-009-0210-z. Epub 2009 Apr 3. Chromosoma. 2009. PMID: 19340450 Free PMC article. Review.
Cited by
- Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation.
Graf R, Munschauer M, Mastrobuoni G, Mayr F, Heinemann U, Kempa S, Rajewsky N, Landthaler M. Graf R, et al. RNA Biol. 2013 Jul;10(7):1146-59. doi: 10.4161/rna.25194. Epub 2013 May 29. RNA Biol. 2013. PMID: 23770886 Free PMC article. - Fibroblast growth factor (FGF) signaling during gastrulation negatively modulates the abundance of microRNAs that regulate proteins required for cell migration and embryo patterning.
Bobbs AS, Saarela AV, Yatskievych TA, Antin PB. Bobbs AS, et al. J Biol Chem. 2012 Nov 9;287(46):38505-14. doi: 10.1074/jbc.M112.400598. Epub 2012 Sep 20. J Biol Chem. 2012. PMID: 22995917 Free PMC article. - Transcriptome-wide analysis of small RNA expression in early zebrafish development.
Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Wei C, et al. RNA. 2012 May;18(5):915-29. doi: 10.1261/rna.029090.111. Epub 2012 Mar 8. RNA. 2012. PMID: 22408181 Free PMC article. - Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii.
Lou S, Sun T, Li H, Hu Z. Lou S, et al. Biotechnol Biofuels. 2018 Sep 8;11:244. doi: 10.1186/s13068-018-1249-y. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30202439 Free PMC article. Review. - Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation.
Wu D, Muhlrad D, Bowler MW, Jiang S, Liu Z, Parker R, Song H. Wu D, et al. Cell Res. 2014 Feb;24(2):233-46. doi: 10.1038/cr.2013.152. Epub 2013 Nov 19. Cell Res. 2014. PMID: 24247251 Free PMC article.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. - PubMed
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- A11832/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 6934/CRUK_/Cancer Research UK/United Kingdom
- A6934/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials