Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene - PubMed (original) (raw)
Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene
Bo Thomsen et al. Neurogenetics. 2010 May.
Abstract
Bovine spinal dysmyelination (BSD) is a recessive congenital neurodegenerative disease in cattle (Bos taurus) characterized by pathological changes of the myelin sheaths in the spinal cord. The occurrence of BSD is a longstanding problem in the American Brown Swiss (ABS) breed and in several European cattle breeds upgraded with ABS. Here, we show that the disease locus on bovine chromosome 11 harbors the SPAST gene that, when mutated, is responsible for the human disorder hereditary spastic paraplegia (HSP). Initially, SPAST encoding Spastin was considered a less likely candidate gene for BSD since the modes of inheritance as well as the time of onset and severity of symptoms differ widely between HSP and BSD. However, sequence analysis of the bovine SPAST gene in affected animals identified a R560Q substitution at a position in the ATPase domain of the Spastin protein that is invariant from insects to mammals. Interestingly, three different mutations in human SPAST gene at the equivalent position are known to cause HSP. To explore this observation further, we genotyped more than 3,100 animals of various cattle breeds and found that the glutamine allele exclusively occurred in breeds upgraded with ABS. Furthermore, all confirmed BSD carriers were heterozygous, while all affected calves were homozygous for the glutamine allele consistent with recessive transmission of the underlying mutation and complete penetrance in the homozygous state. Subsequent analysis of recombinant Spastin in vitro showed that the R560Q substitution severely impaired the ATPase activity, demonstrating a causal relationship between the SPAST mutation and BSD.
Figures
Fig. 1
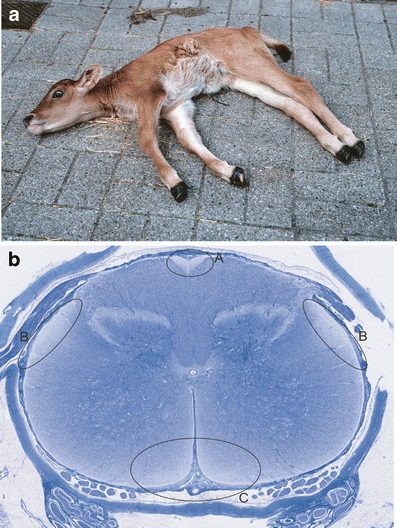
Clinical and pathological characteristics of bovine spinal dysmyelination. a Phenotypic appearance of a calf affected with bovine spinal dysmyelination. b Whole slide digital microscopic image of cervical spinal cord stained with Luxol fast blue. Lack of myelin is seen as reduced blue staining in the ascending gracile funiculus (a), the ascending dorsolateral spinocerebellar tracts (b), and the descending sulcomarginal tracts (c)
Fig. 2
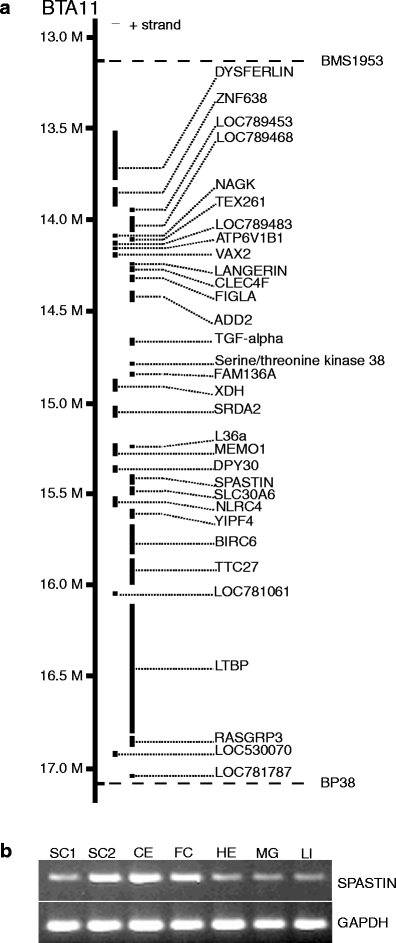
a Physical map of the bovine spinal dysmyelination locus between the linked microsatellite markers BP38 and BMS1953. The location and order of the genes are shown according to the cattle genome assembly Btau_4.0. b Expression analysis by reverse transcription-polymerase chain reaction of SPAST. Tissues analyzed are SC1 spinal cord 1 (cervical), SC2 spinal cord 2 (lumbar), CE cerebellum, FC frontal cortex, HE heart, MG mammary gland, and LI liver
Fig. 3
Multiple sequence alignment of Spastin homologues. Key domains in Spastin are color marked: The AAA domain ATPase associated with diverse cellular activities (yellow), MIT microtubule interacting and trafficking domain (purple), Walker A and B motifs (green), and arginine finger and second region of homology (blue). The arginine residue at position 560 is evolutionary conserved from mammals to insects (red)
Fig. 4
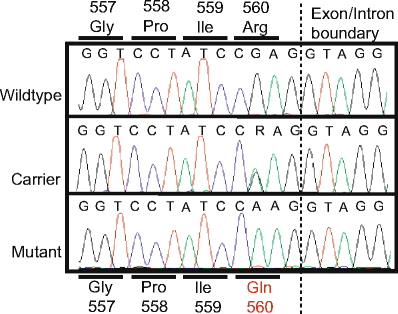
Sequence of the missense mutation causing the R560Q substitution. The chromatograms show the sequence of a genomic fragment spanning the putative disease-causing mutation. Wild-type and affected individuals were homozygous for arginine and glutamine, respectively, whereas individuals with bovine spinal dysmyelination carrier status were heterozygous
Fig. 5
a Coomassie blue stain of a sodium dodecyl sulfate-polyacrylamide gel. Recombinant wild-type and mutant Spastin fused to glutathione _S_-transferase (GST) were expressed in Escherichia coli and purified by affinity chromatography using glutathione sepharose columns. Lane 1, molecular size marker. Lane 2, wild-type GST-Spastin fusion protein. Lane 3, mutant GST-Spastin protein. b ATPase activity of wild-type and mutated Spastin were measured as a function of time. ATP hydrolysis was measured spectrophotometrically using a coupled enzyme assay. Wild-type Spastin denatured by boiling served as a control
Similar articles
- SPAST mutations in Australian patients with hereditary spastic paraplegia.
Vandebona H, Kerr NP, Liang C, Sue CM. Vandebona H, et al. Intern Med J. 2012 Dec;42(12):1342-7. doi: 10.1111/j.1445-5994.2012.02941.x. Intern Med J. 2012. PMID: 23252998 - Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia.
Shoukier M, Neesen J, Sauter SM, Argyriou L, Doerwald N, Pantakani DV, Mannan AU. Shoukier M, et al. Eur J Hum Genet. 2009 Feb;17(2):187-94. doi: 10.1038/ejhg.2008.147. Epub 2008 Aug 13. Eur J Hum Genet. 2009. PMID: 18701882 Free PMC article. - Spastin mutation screening in Chinese patients with pure hereditary spastic paraplegia.
Wei QQ, Chen Y, Zheng ZZ, Chen X, Huang R, Yang Y, Burgunder J, Shang HF. Wei QQ, et al. Parkinsonism Relat Disord. 2014 Aug;20(8):845-9. doi: 10.1016/j.parkreldis.2014.04.021. Epub 2014 May 2. Parkinsonism Relat Disord. 2014. PMID: 24824479 - Detection of novel mutations and review of published data suggests that hereditary spastic paraplegia caused by spastin (SPAST) mutations is found more often in males.
Proukakis C, Moore D, Labrum R, Wood NW, Houlden H. Proukakis C, et al. J Neurol Sci. 2011 Jul 15;306(1-2):62-5. doi: 10.1016/j.jns.2011.03.043. Epub 2011 May 5. J Neurol Sci. 2011. PMID: 21546041 Review. - Hereditary spastic paraplegia SPG4: what is known and not known about the disease.
Solowska JM, Baas PW. Solowska JM, et al. Brain. 2015 Sep;138(Pt 9):2471-84. doi: 10.1093/brain/awv178. Epub 2015 Jun 20. Brain. 2015. PMID: 26094131 Free PMC article. Review.
Cited by
- Management of lethal recessive alleles in beef cattle through the use of mate selection software.
Upperman LR, Kinghorn BP, MacNeil MD, Van Eenennaam AL. Upperman LR, et al. Genet Sel Evol. 2019 Aug 6;51(1):36. doi: 10.1186/s12711-019-0477-3. Genet Sel Evol. 2019. PMID: 31382878 Free PMC article. - Clinicopathological and pedigree investigation of a novel spinocerebellar neurological disease in juvenile Quarter Horses in North America.
Willis AT, Dahlgren AR, Woolard KD, Ghosh S, Donnelly CG, de la Concha-Bermejillo A, Pacheco A, Watson KD, Berryhill E, Aleman M, Wensley F, Humphreys S, Whitehead AE, Goldsmith D, Chesen B, Ragsdale J, Tompkins JE, Nash R, Plunkett AH, Qualls HJ, Rodriguez K, Hochanadel D, Miller AD, Finno CJ. Willis AT, et al. J Vet Intern Med. 2024 May-Jun;38(3):1808-1814. doi: 10.1111/jvim.17049. Epub 2024 Apr 26. J Vet Intern Med. 2024. PMID: 38669583 Free PMC article. - Cross-population selection signatures in Canchim composite beef cattle.
Duarte INH, Bessa AFO, Rola LD, Genuíno MVH, Rocha IM, Marcondes CR, Regitano LCA, Munari DP, Berry DP, Buzanskas ME. Duarte INH, et al. PLoS One. 2022 Apr 1;17(4):e0264279. doi: 10.1371/journal.pone.0264279. eCollection 2022. PLoS One. 2022. PMID: 35363779 Free PMC article. - Confirmation of a non-synonymous SNP in PNPLA8 as a candidate causal mutation for Weaver syndrome in Brown Swiss cattle.
Kunz E, Rothammer S, Pausch H, Schwarzenbacher H, Seefried FR, Matiasek K, Seichter D, Russ I, Fries R, Medugorac I. Kunz E, et al. Genet Sel Evol. 2016 Mar 18;48:21. doi: 10.1186/s12711-016-0201-5. Genet Sel Evol. 2016. PMID: 26992691 Free PMC article. - Screening genetic diseases prevalence in Braunvieh cattle.
Zepeda-Batista JL, Parra-Bracamonte GM, Núñez-Domínguez R, Ramírez-Valverde R, Ruíz-Flores A. Zepeda-Batista JL, et al. Trop Anim Health Prod. 2019 Jan;51(1):25-31. doi: 10.1007/s11250-018-1655-y. Epub 2018 Jul 17. Trop Anim Health Prod. 2019. PMID: 30014197
References
- Charlier C, Coppieters W, Rollin F, Desmecht D, Agerholm JS, Cambisano N, Carta E, Dardano S, Dive M, Fasquelle C, Frennet JC, Hanset R, Hubin X, Jorgensen C, Karim L, Kent M, Harvey K, Pearce BR, Simon P, Tama N, Nie H, Vandeputte S, Lien S, Longeri M, Fredholm M, Harvey RJ, Georges M. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat Genet. 2008;40:449–454. doi: 10.1038/ng.96. - DOI - PubMed
- Agerholm JS, Bendixen C, Andersen O, Arnbjerg J. Complex vertebral malformation in holstein calves. J Vet Diagn Invest. 2001;13:283–289. - PubMed
- Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm LE, Nielsen VH, Agerholm JS, Arnbjerg J, Bendixen C. A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. 2006;16:97–105. doi: 10.1101/gr.3690506. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources