RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells - PubMed (original) (raw)
. 2009 Aug 28;35(4):511-22.
doi: 10.1016/j.molcel.2009.08.002.
Camilla Hauge, Scott R Frank, Claus J Jensen, Katarzyna Duda, Jakob V Nielsen, Michael S Cohen, Jens V Johansen, Benny R Winther, Leif R Lund, Ole Winther, Jack Taunton, Steen H Hansen, Morten Frödin
Affiliations
- PMID: 19716794
- PMCID: PMC3784321
- DOI: 10.1016/j.molcel.2009.08.002
RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells
Ulrik Doehn et al. Mol Cell. 2009.
Abstract
The RAS-stimulated RAF-MEK-ERK pathway confers epithelial cells with critical motile and invasive capacities during development, tissue regeneration, and carcinoma progression, often via promoting the epithelial-mesenchymal transition (EMT). Many mechanisms by which ERK exerts this control remain elusive. We demonstrate that the ERK-activated kinase RSK is necessary to induce mesenchymal motility and invasive capacities in nontransformed epithelial and carcinoma cells. RSK is sufficient to induce certain motile responses. Expression profiling analysis revealed that a primary role of RSK is to induce transcription of a potent promotile/invasive gene program by FRA1-dependent and -independent mechanisms. The program enables RSK to coordinately modulate the extracellular environment, the intracellular motility apparatus, and receptors mediating communication between these compartments to stimulate motility and invasion. These findings uncover a mechanism whereby the RAS-ERK pathway controls epithelial cell motility by identifying RSK as a key effector, from which emanate multiple highly coordinate transcription-dependent mechanisms for stimulation of motility and invasive properties.
Figures
Figure 1. RSK is a necessary, and partially sufficient, effector of the RAS-ERK pathway for induction of a motile, mesenchymal phenotype in epithelial cells
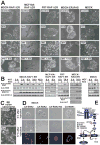
(A) Native or RAF1:ER- or RAS:ER-expressing epithelial cell lines were exposed to 1 μM 4HT, 30 nM hepatocyte growth factor (HGF) or 10 μM fmk as indicated, and photographed after 24–48 h. (B) Cells treated as in (A) were analysed by immunoblotting. (C) BE cells were cultured in the absence or presence of fmk and photographed after 48 h. (D) MDCK cells microinjected with plasmids expressing epitope-tagged, constitutively active (CA) or kinase-dead (KD) mutants of ERK2, RSK2 or MSK1 were photographed 0 h or 17 h after injection and thereafter subjected to immunostaining against the epitope tag. (E) The RAS-activated RAF-MEK-ERK-RSK pathway and sites of action of kinase inhibitors used here. See Fig. S1 for details on RSK structure. Experiments were conducted 3–6 times with similar results.
Figure 2. RSK is required for several forms of ERK-stimulated motility in diverse immortalized or cancerous epithelial cell types
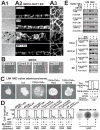
(A) Confluent, polarized MDCK-RAF1:ER cells on filters were exposed to 1 μM 4HT or 10 μM fmk as indicated. After 24 h, the cells were analysed by phase contrast microscopy (arrows indicate areas with 2–4 cell thick multilayering) (A1) or by actin filament staining and confocal microscopy in Z-X (A2) sections or X-Y sections at the base of the cell layer (A3). (B) MDCK cell monolayers were wounded in the absence or presence of 10 μM fmk and photographed at 0 h or 24 h. (C) 3D organoids of LIM 1863 colon adenocarcinoma cells were exposed to 10 ng/ml TNFα, 2 ng/ml TGFβ, 10 μM fmk and 10 μM U0126 as indicated, and photographed after 24 h or analysed for chemotactic cell migration in Boyden chamber assays, using NIH-3T3 cell conditioned medium (CM) as chemoattractant. (D) Various immortalized or transformed epithelial cell lines were treated, or not, for 24 h with 1 μM 4HT, 6 μM fmk or 10 μM BI-D1870 as indicated and then subjected to 3D invasion ssay through Matrigel basement membrane matrix, using indicated chemoattractants. After 24 h, cells that had invaded through the matrix were quantified and expressed as percent of maximum. Photographs of filters with invaded, crystal violet-stained cells from representative experiments are shown here and in Fig. S2. (E) Cells treated as in (C) and (D) were analysed by immunoblotting. pS1798 blotting was performed on immunopurified TSC2. Experiments were conducted 3–6 times with similar results. Data in (C) and (D) are mean ±SD of 3 experiments.
Figure 3. RSK induces the expression of a coordinate motility and invasion gene program in MDCK cells
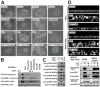
(A) Polarized MDCK-RAF1:ER cell monolayers were exposed to 1 μM 4HT and 6 μM fmk as indicated. After 24 h, the cells or the medium were analyzed by immunoblotting. (B) Subconfluent MDCK cells were cultured in the absence or presence of 25 μM laminin 332 and photographed 48 h later. (C) Polarized MDCK-RAF1:ER cell monolayers treated as in (A) were analyzed for active Rac1. All experiments were conducted 3–5 times with similar results.
Figure 4. Effects of a rigorous RSK inhibition protocol and an fmk-resistant RSK2 mutant on MDCK cell motility and expression of pro-motile/invasive proteins
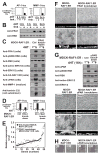
(A) MDCK-RAF1:ER cells were subjected to 6 μM fmk pulse (1 h fmk incubation followed by extensive washing), 10 μM BI-D1870, 100 μM SL0101, 5 μM GF109203X and 1 μM 4HT as indicated, and photographed after 24 h. (B) Lysates or medium from cells treated as in (A) were analysed by immunoblotting. (C) MDCK-RAF1:ER cells transfected with plasmid expressing HA-tagged RSK1, -2, -3 or -4 were exposed to 6 μM fmk and 1 μM 4HT, as indicated. After 24 h, the cells were analyzed by immunoblotting. (D) MDCK-RAF1:ER cells stably expressing wild-type RSK2 or fmk-resistant RSK2-C436V mutant were analyzed in multilayering assays or by immunoblotting. Experiments were conducted 3 times with similar results.
Figure 5. RSK induces the expression of FRA1 to promote induction of pro-motile/invasive genes and phenotypes in MDCK epithelial cells
(A) MDCK-RAF1:ER cells transfected with AP1-luc or MMP1-luc reporter plasmids were exposed to 1 μM 4HT and 6 μM fmk as indicated, and analysed for reporter gene expression after 24 h. (B) MDCK-RAF1:ER cells were exposed to 1 μM 4HT and 6 μM fmk as indicated, and analysed by immunoblotting. (C) MDCK-RAF1:ER cells without (Ctrl) or with knockdown of FRA1 were exposed to 1 μM 4HT as indicated, and analysed by immunoblotting after 24 h. (D) MDCK-RAF1:ER cells without (Ctrl) or with knockdown of FRA1 were transfected with AP1-luc or MMP1-luc reporter plasmids and analysed as described in (B). (E) Graphical representation illustrating that the 53 fmk-sensitive genes that were also sensitive to FRA1 knockdown, quantitatively shows very similar sensitivity. Dot and diamond located at the same position of the X axis represent the same gene. (F) MDCK-RAF1:ER cells with or without knockdown of FRA1 were exposed or not to 1 μM 4HT as indicated, and assessed for multilayering after 24 h, as described in legend to Fig. 2A. (G) MDCK-RAF1:ER cells without (Ctrl) or with uPAR knockdown were treated and analyzed as described in (C). (H) Wound healing assays on MDCK-RAF1:ER cells without (Ctrl) or with uPAR knockdown were performed as described in Fig. 2B. Experiments were conducted 3–4 times with similar results. Data in (A) and (D) are mean ±SD of 3 independent experiments.
Figure 6. RSK stimulates a pro-motile/invasive gene program in various non-transformed or cancerous epithelial cell types
MCF10A-RAF1:ER (A), MCF10A (B) and LIM 1863 cells (C) were exposed to 1 μM 4HT, 20 nM EGF, 10 ng/ml TNFα, 2 ng/ml TGFβ, 10 μM BI-D1870, 6 μM fmk or 10 μM U0126, as indicated. After 24 h, the cells or medium were analyzed by immunoblotting. (D) MDCK cells expressing conditionally active CA-RSK2 or vector Ctrl were exposed or not to 1 μM of the inducer Shield1. The cells were analysed after 24 h by immunoblotting or assessed for multilayering after 72 h, as described in legend to Fig. 2A. (E) MCF10A-RAF1:ER cells were subjected to siRNA knockdown of RSK1-4 in the combinations indicated. After 48 h, the cells were analysed by immunoblotting or in invasion assays, as described in the legend to Fig. 2D. (F) The model summarizes the results of the present study and illustrates that RSK may induce mesenchymal invasive capacities in epithelial cells by stimulating a coordinate gene program (green) via FRA1-dependent and -independent mechanisms.
Similar articles
- ERK and RSK regulate distinct steps of a cellular program that induces transition from multicellular epithelium to single cell phenotype.
Čáslavský J, Klímová Z, Vomastek T. Čáslavský J, et al. Cell Signal. 2013 Dec;25(12):2743-51. doi: 10.1016/j.cellsig.2013.08.024. Epub 2013 Sep 3. Cell Signal. 2013. PMID: 24012955 - The N-terminal region of p27 inhibits HIF-1α protein translation in ribosomal protein S6-dependent manner by regulating PHLPP-Ras-ERK-p90RSK axis.
Zhang D, Liu J, Mi X, Liang Y, Li J, Huang C. Zhang D, et al. Cell Death Dis. 2014 Nov 20;5(11):e1535. doi: 10.1038/cddis.2014.496. Cell Death Dis. 2014. PMID: 25412313 Free PMC article. - Sustained c-Jun-NH2-kinase activity promotes epithelial-mesenchymal transition, invasion, and survival of breast cancer cells by regulating extracellular signal-regulated kinase activation.
Wang J, Kuiatse I, Lee AV, Pan J, Giuliano A, Cui X. Wang J, et al. Mol Cancer Res. 2010 Feb;8(2):266-77. doi: 10.1158/1541-7786.MCR-09-0221. Epub 2010 Feb 9. Mol Cancer Res. 2010. PMID: 20145041 Free PMC article. - The RSK factors of activating the Ras/MAPK signaling cascade.
Carriere A, Ray H, Blenis J, Roux PP. Carriere A, et al. Front Biosci. 2008 May 1;13:4258-75. doi: 10.2741/3003. Front Biosci. 2008. PMID: 18508509 Review. - RSK isoforms in cancer cell invasion and metastasis.
Sulzmaier FJ, Ramos JW. Sulzmaier FJ, et al. Cancer Res. 2013 Oct 15;73(20):6099-105. doi: 10.1158/0008-5472.CAN-13-1087. Epub 2013 Oct 4. Cancer Res. 2013. PMID: 24097826 Free PMC article. Review.
Cited by
- Role of epidermal growth factor receptor in breast cancer.
Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Masuda H, et al. Breast Cancer Res Treat. 2012 Nov;136(2):331-45. doi: 10.1007/s10549-012-2289-9. Epub 2012 Oct 17. Breast Cancer Res Treat. 2012. PMID: 23073759 Free PMC article. Review. - Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Serafimova IM, Pufall MA, Krishnan S, Duda K, Cohen MS, Maglathlin RL, McFarland JM, Miller RM, Frödin M, Taunton J. Serafimova IM, et al. Nat Chem Biol. 2012 Apr 1;8(5):471-6. doi: 10.1038/nchembio.925. Nat Chem Biol. 2012. PMID: 22466421 Free PMC article. - The role of the p90 ribosomal S6 kinase family in prostate cancer progression and therapy resistance.
Cronin R, Brooke GN, Prischi F. Cronin R, et al. Oncogene. 2021 Jun;40(22):3775-3785. doi: 10.1038/s41388-021-01810-9. Epub 2021 May 10. Oncogene. 2021. PMID: 33972681 Free PMC article. Review. - PDGFRα signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways.
Clement DL, Mally S, Stock C, Lethan M, Satir P, Schwab A, Pedersen SF, Christensen ST. Clement DL, et al. J Cell Sci. 2013 Feb 15;126(Pt 4):953-65. doi: 10.1242/jcs.116426. Epub 2012 Dec 21. J Cell Sci. 2013. PMID: 23264740 Free PMC article. - miRNA Alterations Modify Kinase Activation In The IGF-1 Pathway And Correlate With Colorectal Cancer Stage And Progression In Patients.
Knowlton DL, Tang K, Henstock PV, Subramanian RR. Knowlton DL, et al. J Cancer. 2011;2:490-502. doi: 10.7150/jca.2.490. Epub 2011 Oct 1. J Cancer. 2011. PMID: 21980324 Free PMC article.
References
- Bates RC, Goldsmith JD, Bachelder RE, Brown C, Shibuya M, Oettgen P, Mercurio AM. Flt-1-dependent survival characterizes the epithelial-mesenchymal transition of colonic organoids. Curr Biol. 2003;13:1721–1727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071434/GM/NIGMS NIH HHS/United States
- R01 CA092354/CA/NCI NIH HHS/United States
- U18 DP006133/DP/NCCDPHP CDC HHS/United States
- U18DP006133/ACL/ACL HHS/United States
- R01 CA092354-05/CA/NCI NIH HHS/United States
- R01 GM71434/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous