Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta - PubMed (original) (raw)
Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta
Michael K Swan et al. Nat Struct Mol Biol. 2009 Sep.
Abstract
DNA polymerase delta (Pol delta) is a high-fidelity polymerase that has a central role in replication from yeast to humans. We present the crystal structure of the catalytic subunit of yeast Pol delta in ternary complex with a template primer and an incoming nucleotide. The structure, determined at 2.0-A resolution, catches the enzyme in the act of replication, revealing how the polymerase and exonuclease domains are juxtaposed relative to each other and how a correct nucleotide is selected and incorporated. The structure also reveals the 'sensing' interactions near the primer terminus, which signal a switch from the polymerizing to the editing mode. Taken together, the structure provides a chemical basis for the bulk of DNA synthesis in eukaryotic cells and a framework for understanding the effects of cancer-causing mutations in Pol delta.
Figures
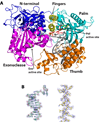
Figure 1. Structure of the Pol3-DNA-dCTP ternary complex
(A) The Pol3 palm, fingers, thumb, exonuclease, and the N-terminal domains are shown in cyan, yellow, orange, magenta and dark blue, respectively. DNA is in gray, template G and incoming dCTP are in red, and the Ca2+ ions are in green. The polymerase (Pol) and exonuclease (Exo) active sites are marked, and the residues that comprise the two active sites are highlighted. B) Experimental electron density at 2.7Ǻ after solvent flattening, around the 12/16 primer-template (left) and helix P (right; c.f. Supplementary Fig. 1). The density is contoured at 1.0σ.
Figure 2. Comparison between Pol3 and RB69 Pol ternary complexes
(A) Pol3 and RB69 ternary complexes displayed in the same orientation, based on a superposition of their palm domains. The color scheme is the same as in Fig.1, where the palm, fingers, thumb, exonuclease, and the N-terminal domains are shown in cyan, yellow, orange, magenta and dark blue, respectively. DNA is in gray; template G (Pol3) and A (RB69), and incoming dCTP (Pol3) and dTTP (RB69) are in red; Ca2+ ions (Pol3 and Rb69) are in green. The polymerase (Pol) and exonuclease (Exo) active sites are marked, and the residues that comprise the two active sites are highlighted in stick form. (B) The Pol3 surface in two orientations. Highlighted in green are the sites of identical residues between Pol3 and Rb69 Pol (c.f. Supplementary Fig. 1), and highlighted in red is the DNA in the two orientations. Also highlighted, alongside the figures, are the “local” structure based sequence alignments between Pol3 (upper) and RB69 Pol (lower) (c.f. Supplementary Fig. 1) over the green “patches” of sequence identity. Note that the most prominent patches of sequence identity are centered over the Pol active site and the nascent base pair, the Exo active site, and the minor groove “sensing” residues.
Figure 3. Close-up view of the polymerase active site region
The fingers, palm, exonuclease, and the N-terminal domains are colored in faded yellow, cyan, magenta, and blue. The DNA is colored gray, templating G and incoming dCTP are in red, and the Ca2+ ions (A, B and C) are in green. Highlighted and labeled are the acidic residues (Asp608, Asp764 and Glu802); residues that interact with the triphosphate moiety of incoming dCTP via side chain (Arg674 and Lys701), main chain (Ser611 and Leu612), and a water molecule (Lys678); residues that impinge on the dG-dCTP bases (Asn705, Ser706, Tyr708, and Glu709); and Tyr613 that stacks against the dCTP sugar. The residues are colored to match the domain they belong to.
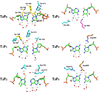
Figure 4. Pol3-DNA base interactions
The bases, at the replicative end of the template-primer, are labeled Tn-Pn (where T and P refer to template and primer strands, respectively, and the subscripts n (0, 1, 2 and 3) refer to the number of base pairs from the templating base position). The Pol3 residues are colored to match the domain they belong to: yellow for fingers, cyan for palm, and magenta for exonuclease. The water molecules are colored red. Dashed lines depict hydrogen bonds (with distances in Angstroms above the bonds).
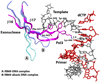
Figure 5. β-hairpin-DNA interactions
In Pol3, an extended β-hairpin on the exonuclease domain inserts into the DNA major groove and interacts extensively with the unpaired portion of the template strand. Also shown for comparison are the β-hairpin positions in RB69 Pol complexed with normal DNA (A) or with DNA containing an abasic residue at the templating site (B).
Figure 6. Mapping of cancer mutations
Pol3 is the site of several mutations that accelerate tumorigenesis in mice and humans. The equivalent residues in yeast Pol3 (Asp407, Arg511, Leu612, Arg658, Arg696 and Lys753) are mapped onto the ternary complex to show their relative locations with respect to the polymerase (Pol) and exonuclease (Exo) active sites.
Similar articles
- DNA polymerase 3'→5' exonuclease activity: Different roles of the beta hairpin structure in family-B DNA polymerases.
Darmawan H, Harrison M, Reha-Krantz LJ. Darmawan H, et al. DNA Repair (Amst). 2015 May;29:36-46. doi: 10.1016/j.dnarep.2015.02.014. Epub 2015 Feb 23. DNA Repair (Amst). 2015. PMID: 25753811 - Cryo-EM structure and dynamics of eukaryotic DNA polymerase δ holoenzyme.
Jain R, Rice WJ, Malik R, Johnson RE, Prakash L, Prakash S, Ubarretxena-Belandia I, Aggarwal AK. Jain R, et al. Nat Struct Mol Biol. 2019 Oct;26(10):955-962. doi: 10.1038/s41594-019-0305-z. Epub 2019 Oct 3. Nat Struct Mol Biol. 2019. PMID: 31582849 Free PMC article. - Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA.
Zheng F, Georgescu RE, Li H, O'Donnell ME. Zheng F, et al. Proc Natl Acad Sci U S A. 2020 Dec 1;117(48):30344-30353. doi: 10.1073/pnas.2017637117. Epub 2020 Nov 17. Proc Natl Acad Sci U S A. 2020. PMID: 33203675 Free PMC article. - Structure and function of eukaryotic DNA polymerase δ.
Tahirov TH. Tahirov TH. Subcell Biochem. 2012;62:217-36. doi: 10.1007/978-94-007-4572-8_12. Subcell Biochem. 2012. PMID: 22918588 Review. - DNA polymerase delta, an essential enzyme for DNA transactions.
Hindges R, Hübscher U. Hindges R, et al. Biol Chem. 1997 May;378(5):345-62. doi: 10.1515/bchm.1997.378.5.345. Biol Chem. 1997. PMID: 9191022 Review.
Cited by
- PCNA trimer instability inhibits translesion synthesis by DNA polymerase η and by DNA polymerase δ.
Dieckman LM, Washington MT. Dieckman LM, et al. DNA Repair (Amst). 2013 May 1;12(5):367-76. doi: 10.1016/j.dnarep.2013.02.007. Epub 2013 Mar 15. DNA Repair (Amst). 2013. PMID: 23506842 Free PMC article. - A Major Role of DNA Polymerase δ in Replication of Both the Leading and Lagging DNA Strands.
Johnson RE, Klassen R, Prakash L, Prakash S. Johnson RE, et al. Mol Cell. 2015 Jul 16;59(2):163-175. doi: 10.1016/j.molcel.2015.05.038. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145172 Free PMC article. - Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas.
Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S; CORGI Consortium; WGS500 Consortium; Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I. Palles C, et al. Nat Genet. 2013 Feb;45(2):136-44. doi: 10.1038/ng.2503. Epub 2012 Dec 23. Nat Genet. 2013. PMID: 23263490 Free PMC article. - Beyond the Lesion: Back to High Fidelity DNA Synthesis.
Kaszubowski JD, Trakselis MA. Kaszubowski JD, et al. Front Mol Biosci. 2022 Jan 5;8:811540. doi: 10.3389/fmolb.2021.811540. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071328 Free PMC article. Review. - Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant.
Xia S, Konigsberg WH. Xia S, et al. Protein Sci. 2014 Apr;23(4):508-13. doi: 10.1002/pro.2434. Protein Sci. 2014. PMID: 24458997 Free PMC article.
References
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. - PubMed
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. - PubMed
- Hartwell LH. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases