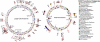Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae - PubMed (original) (raw)
Comparative Study
. 2009 Sep 8;106(36):15442-7.
doi: 10.1073/pnas.0907787106. Epub 2009 Aug 31.
Christopher J Grim, Nur A Hasan, Je Hee Lee, Seon Young Choi, Bradd J Haley, Elisa Taviani, Yoon-Seong Jeon, Dong Wook Kim, Jae-Hak Lee, Thomas S Brettin, David C Bruce, Jean F Challacombe, J Chris Detter, Cliff S Han, A Christine Munk, Olga Chertkov, Linda Meincke, Elizabeth Saunders, Ronald A Walters, Anwar Huq, G Balakrish Nair, Rita R Colwell
Affiliations
- PMID: 19720995
- PMCID: PMC2741270
- DOI: 10.1073/pnas.0907787106
Comparative Study
Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae
Jongsik Chun et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Vibrio cholerae, the causative agent of cholera, is a bacterium autochthonous to the aquatic environment, and a serious public health threat. V. cholerae serogroup O1 is responsible for the previous two cholera pandemics, in which classical and El Tor biotypes were dominant in the sixth and the current seventh pandemics, respectively. Cholera researchers continually face newly emerging and reemerging pathogenic clones carrying diverse combinations of phenotypic and genotypic properties, which significantly hampered control of the disease. To elucidate evolutionary mechanisms governing genetic diversity of pandemic V. cholerae, we compared the genome sequences of 23 V. cholerae strains isolated from a variety of sources over the past 98 years. The genome-based phylogeny revealed 12 distinct V. cholerae lineages, of which one comprises both O1 classical and El Tor biotypes. All seventh pandemic clones share nearly identical gene content. Using analogy to influenza virology, we define the transition from sixth to seventh pandemic strains as a "shift" between pathogenic clones belonging to the same O1 serogroup, but from significantly different phyletic lineages. In contrast, transition among clones during the present pandemic period is characterized as a "drift" between clones, differentiated mainly by varying composition of laterally transferred genomic islands, resulting in emergence of variants, exemplified by V. cholerae O139 and V. cholerae O1 El Tor hybrid clones. Based on the comparative genomics it is concluded that V. cholerae undergoes extensive genetic recombination via lateral gene transfer, and, therefore, genome assortment, not serogroup, should be used to define pathogenic V. cholerae clones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
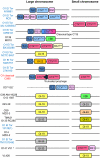
Fig. 1.
Neighbor-joining trees showing phylogenetic relationships of 23 V. cholerae strains representing diverse serogroups. (A) All V. cholerae strains based on 1,676 genes (1,370,469 bp). (B) Phylocore genome (PG) clade based on 2,663 genes (2,567,393 bp). (C) Seventh pandemic (7P) clade based on 3,364 genes (3,291,577 bp). Bootstrap supports, as percentage, are indicated at the branching points. Bars represent the numbers of substitution per site, respectively. Only orthologous genes showing >95% nucleotide sequence similarity to those of V. cholerae N16961 were selected. The tree was rooted using Vibrio vulnificus YJ016 and Vibrio parahaemolyticus RIMD 2210633.
Fig. 2.
Schematic representation of various prophages and genetic elements present in the target regions of CTXφ insetion. *, TLC, El Tor type CTXφ, RS1 element are found, but no positional information can be obtained from genome assemblies. †, classical type CTXφ and RS1 are present, but no positional information can be obtained.
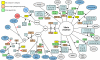
Fig. 3.
Genomic representation of genomic islands of both V. cholerae chromosomes. The two circles in the middle represent the genes in V. cholerae O1 El Tor N16961. The inner circle indicates genomic islands found in strain N16961, whereas the outer circles are those absent in strain N16961.
Fig. 4.
Proposed hypothetical evolutionary pathway of the V. cholerae species. Probable insertions and deletions of genomic islands (
Table S1
) found in 23 V. cholerae strains are indicated by black and red arrows, respectively, along the phylogenetic tree based on genome sequence data. Hypothetical ancestral strains are indicated by open circles.
Similar articles
- Novel Cholera Toxin Variant and ToxT Regulon in Environmental Vibrio mimicus Isolates: Potential Resources for the Evolution of Vibrio cholerae Hybrid Strains.
Neogi SB, Chowdhury N, Awasthi SP, Asakura M, Okuno K, Mahmud ZH, Islam MS, Hinenoya A, Nair GB, Yamasaki S. Neogi SB, et al. Appl Environ Microbiol. 2019 Jan 23;85(3):e01977-18. doi: 10.1128/AEM.01977-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30446560 Free PMC article. - Comparative genomics for non-O1/O139 Vibrio cholerae isolates recovered from the Yangtze River Estuary versus V. cholerae representative isolates from serogroup O1.
Gong L, Yu P, Zheng H, Gu W, He W, Tang Y, Wang Y, Dong Y, Peng X, She Q, Xie L, Chen L. Gong L, et al. Mol Genet Genomics. 2019 Apr;294(2):417-430. doi: 10.1007/s00438-018-1514-6. Epub 2018 Nov 28. Mol Genet Genomics. 2019. PMID: 30488322 - Characterization and Genetic Variation of Vibrio cholerae Isolated from Clinical and Environmental Sources in Thailand.
Siriphap A, Leekitcharoenphon P, Kaas RS, Theethakaew C, Aarestrup FM, Sutheinkul O, Hendriksen RS. Siriphap A, et al. PLoS One. 2017 Jan 19;12(1):e0169324. doi: 10.1371/journal.pone.0169324. eCollection 2017. PLoS One. 2017. PMID: 28103259 Free PMC article. - Evolution of new variants of Vibrio cholerae O1.
Safa A, Nair GB, Kong RY. Safa A, et al. Trends Microbiol. 2010 Jan;18(1):46-54. doi: 10.1016/j.tim.2009.10.003. Epub 2009 Nov 26. Trends Microbiol. 2010. PMID: 19942436 Review. - Genomic evolution of Vibrio cholerae.
Cho YJ, Yi H, Lee JH, Kim DW, Chun J. Cho YJ, et al. Curr Opin Microbiol. 2010 Oct;13(5):646-51. doi: 10.1016/j.mib.2010.08.007. Epub 2010 Sep 17. Curr Opin Microbiol. 2010. PMID: 20851041 Review.
Cited by
- Genomic characterization of four Escherichia coli strains isolated from oral lichen planus biopsies.
Min H, Baek K, Lee A, Seok YJ, Choi Y. Min H, et al. J Oral Microbiol. 2021 Mar 29;13(1):1905958. doi: 10.1080/20002297.2021.1905958. J Oral Microbiol. 2021. PMID: 33828821 Free PMC article. - Comparative analysis of mobilizable genomic islands.
Daccord A, Ceccarelli D, Rodrigue S, Burrus V. Daccord A, et al. J Bacteriol. 2013 Feb;195(3):606-14. doi: 10.1128/JB.01985-12. Epub 2012 Nov 30. J Bacteriol. 2013. PMID: 23204461 Free PMC article. - Draft Genome sequence of Escherichia coli AI27, a porcine isolate belonging to phylogenetic group B1.
Lee K, Yi H, Cho YJ, Jang J, Hur HG, Chun J. Lee K, et al. J Bacteriol. 2012 Dec;194(23):6640-1. doi: 10.1128/JB.01749-12. J Bacteriol. 2012. PMID: 23144394 Free PMC article. - Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America.
Lam C, Octavia S, Reeves P, Wang L, Lan R. Lam C, et al. Emerg Infect Dis. 2010 Jul;16(7):1130-2. doi: 10.3201/eid1607.100131. Emerg Infect Dis. 2010. PMID: 20587187 Free PMC article. - Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea.
Caro-Quintero A, Deng J, Auchtung J, Brettar I, Höfle MG, Klappenbach J, Konstantinidis KT. Caro-Quintero A, et al. ISME J. 2011 Jan;5(1):131-40. doi: 10.1038/ismej.2010.93. Epub 2010 Jul 1. ISME J. 2011. PMID: 20596068 Free PMC article.
References
- Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim Biophys Acta. 2003;1639:65–79. - PubMed
- Colwell RR. Global climate and infectious disease: The cholera paradigm. Science. 1996;274:2025–2031. - PubMed
- Ramamurthy T, et al. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous