REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis - PubMed (original) (raw)
REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis
Gwendal Le Martelot et al. PLoS Biol. 2009 Sep.
Abstract
In mammals, many aspects of behavior and physiology, and in particular cellular metabolism, are coordinated by the circadian timing system. Molecular clocks are thought to rely on negative feedback loops in clock gene expression that engender oscillations in the accumulation of transcriptional regulatory proteins, such as the orphan receptor REV-ERBalpha. Circadian transcription factors then drive daily rhythms in the expression of clock-controlled output genes, for example genes encoding enzymes and regulators of cellular metabolism. To gain insight into clock output functions of REV-ERBalpha, we carried out genome-wide transcriptome profiling experiments with liver RNA from wild-type mice, Rev-erbalpha knock-out mice, or REV-ERBalpha overexpressing mice. On the basis of these genetic loss- and gain-of-function experiments, we concluded that REV-ERBalpha participates in the circadian modulation of sterol regulatory element-binding protein (SREBP) activity, and thereby in the daily expression of SREBP target genes involved in cholesterol and lipid metabolism. This control is exerted via the cyclic transcription of Insig2, encoding a trans-membrane protein that sequesters SREBP proteins to the endoplasmic reticulum membranes and thereby interferes with the proteolytic activation of SREBPs in Golgi membranes. REV-ERBalpha also participates in the cyclic expression of cholesterol-7alpha-hydroxylase (CYP7A1), the rate-limiting enzyme in converting cholesterol to bile acids. Our findings suggest that this control acts via the stimulation of LXR nuclear receptors by cyclically produced oxysterols. In conclusion, our study suggests that rhythmic cholesterol and bile acid metabolism is not just driven by alternating feeding-fasting cycles, but also by REV-ERBalpha, a component of the circadian clockwork circuitry.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
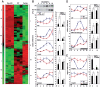
Figure 1. REV-ERBα controls the temporal nuclear accumulation of SREBP and the transcription of SREBP target genes.
(A) Affymetrix-microarray analysis of liver RNA from mice of various genotypes. The heat map displays transcripts with differential accumulation in WT, Rev-KO, and TgRev mice. Differentially expressed transcripts of mice with different genotypes are clustered into four classes: class I: Rev-KO>WT≥TgRev; class II: Rev-KO>WT
<tgrev; iii:="" rev-ko<wt≤tgrev;="" iv:="" rev-ko<wt="">TgRev (see also Figure S2). (B) Temporal accumulation of SREBP1c in liver nuclear extract. (C) Temporal expression of transcripts from selected SREBP1c genes. (D) Temporal expression of transcripts from selected SREBP2 target genes. (E) Temporal expression of Insig2 transcripts. The data displayed in (C–E) were obtained by quantitative (Q) RT-PCR experiments on whole-cell liver RNA from WT, Rev-KO, and TgRev animals. The data represent the mean±SEM (n = 4–6).</tgrev;>
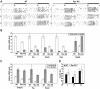
Figure 2. Fasting–feeding regulation of SREBP targeted genes in Rev-KO mice.
(A) Feeding activity of three Rev-KO and WT mice as measured by an infrared detection system. (B) Mice that were either fasted for 24 h or (C) fed a high carbohydrate–no fat diet for 18 d were humanely killed at ZT12, and the accumulation of selected mRNAs was measured in WT and Rev-KO (n = 4 per each data point) using Q-RT PCR. (D) Insulin measurement from blood samples collected at ZT4, ZT12, and ZT16 (n = 5 per each data point). (B–D) data represent the mean±SEM.
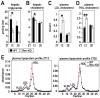
Figure 3. Accumulation of cholesterol and triglycerides in liver and plasma of WT and Rev-KO mice.
(A) Total hepatic triglycerides and (B) cholesterol in WT mice (ZT12 n = 8, ZT20 n = 6) and Rev-KO mice (ZT12 n = 10, ZT20 n = 4). (C) Plasma LDL-cholesterol and (D) HDL-cholesterol in WT and Rev-KO (n = 6 per each data point). (E) Plasma samples from animals humanely killed at ZT12 and ZT20 were pooled (n = 6) and lipoproteins separated by fast protein liquid chromatography (FPLC). (A–D) data represent the mean±SEM. _p_-Values: *, <0.05; **, <0.005.
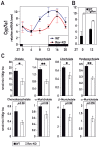
Figure 4. REV-ERBα controls Cyp7a1 transcription and bile acid accumulation.
(A) Temporal Cyp7a1 mRNA accumulation in the livers of WT, Rev-KO, and (B) TgRev mice. The data represent the mean±SEM (n = 4–6). (C) Primary and secondary bile acid levels in gallbladders from WT and Rev-KO mice. Data represent the mean±SEM (n = 6). _p_-Values: *, <0.05; **, <0.005.
Figure 5. LXR participates in the circadian transcription of Cyp7a1.
Temporal Cyp7a1 mRNA accumulation was measured in the livers of (A) Dbp/Tef/Hlf triple knock-out mice (B) E4bp4/Nfil3 knock-out mice and (C) _LXR_α/β double knock-out animals. Data represent the mean±SEM (n = 4).
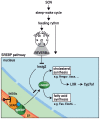
Figure 6. The role of REV-ERBα in circadian lipid and bile acid homeostasis.
The cartoon illustrates how REV-ERBα may modulate circadian lipid, cholesterol, and bile acid synthesis by controlling the accumulation of SREBP in the nucleus. In liver hepatocytes, REV-ERBα accumulates to maximal levels at ZT8–ZT12 and represses Insig2 transcription. Thereby, it promotes the proteolytic activation and nuclear accumulation of SREBP proteins. In turn, the circadian activation of SREBP transcription factors drives the cyclic transcription of Hmgcr, encoding the rate-limiting enzyme of cholesterol biosynthesis. As a consequence the levels of oxysterols, which serve as ligands for LXR, also oscillate during the day, and cyclically activated LXR then controls rhythmic Cyp7a1 transcription.
Similar articles
- Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha.
Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, Wolters H, van der Sluijs FH, Vennström B, Kuipers F, Staels B. Duez H, et al. Gastroenterology. 2008 Aug;135(2):689-98. doi: 10.1053/j.gastro.2008.05.035. Epub 2008 May 15. Gastroenterology. 2008. PMID: 18565334 - Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis.
Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, Kato Y. Noshiro M, et al. J Biol Rhythms. 2007 Aug;22(4):299-311. doi: 10.1177/0748730407302461. J Biol Rhythms. 2007. PMID: 17660447 - REV-ERB_α_ Regulates CYP7A1 Through Repression of Liver Receptor Homolog-1.
Zhang T, Zhao M, Lu D, Wang S, Yu F, Guo L, Wen S, Wu B. Zhang T, et al. Drug Metab Dispos. 2018 Mar;46(3):248-258. doi: 10.1124/dmd.117.078105. Epub 2017 Dec 13. Drug Metab Dispos. 2018. PMID: 29237721 - Dissecting the Rev-erbα Cistrome and the Mechanisms Controlling Circadian Transcription in Liver.
Fang B, Lazar MA. Fang B, et al. Cold Spring Harb Symp Quant Biol. 2015;80:233-8. doi: 10.1101/sqb.2015.80.027508. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370410 Review. - Rev-erbα and the circadian transcriptional regulation of metabolism.
Gerhart-Hines Z, Lazar MA. Gerhart-Hines Z, et al. Diabetes Obes Metab. 2015 Sep;17 Suppl 1(0 1):12-6. doi: 10.1111/dom.12510. Diabetes Obes Metab. 2015. PMID: 26332963 Free PMC article. Review.
Cited by
- Circadian rhythm and atherosclerosis (Review).
Zhang Z, Yu B, Wang X, Luo C, Zhou T, Zheng X, Ding J. Zhang Z, et al. Exp Ther Med. 2020 Nov;20(5):96. doi: 10.3892/etm.2020.9224. Epub 2020 Sep 16. Exp Ther Med. 2020. PMID: 32973945 Free PMC article. Review. - Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages.
Eichenfield DZ, Troutman TD, Link VM, Lam MT, Cho H, Gosselin D, Spann NJ, Lesch HP, Tao J, Muto J, Gallo RL, Evans RM, Glass CK. Eichenfield DZ, et al. Elife. 2016 Jul 27;5:e13024. doi: 10.7554/eLife.13024. Elife. 2016. PMID: 27462873 Free PMC article. - Hypolipidemic effect of Alisma orientale (Sam.) Juzep on gut microecology and liver transcriptome in diabetic rats.
Xu X, Li L, Zhang Y, Lu X, Lin W, Wu S, Qin X, Xu R, Lin W. Xu X, et al. PLoS One. 2020 Oct 9;15(10):e0240616. doi: 10.1371/journal.pone.0240616. eCollection 2020. PLoS One. 2020. PMID: 33035272 Free PMC article. - Metabolic and chemical architecture of the mammalian circadian clock.
Laothamatas I, Rasmussen ES, Green CB, Takahashi JS. Laothamatas I, et al. Cell Chem Biol. 2023 Sep 21;30(9):1033-1052. doi: 10.1016/j.chembiol.2023.08.014. Epub 2023 Sep 13. Cell Chem Biol. 2023. PMID: 37708890 Free PMC article. Review. - Bile acids in glucose metabolism in health and disease.
Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Shapiro H, et al. J Exp Med. 2018 Feb 5;215(2):383-396. doi: 10.1084/jem.20171965. Epub 2018 Jan 16. J Exp Med. 2018. PMID: 29339445 Free PMC article. Review.
References
- Gachon F, Nagoshi E, Brown S. A, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. - PubMed
- Reppert S. M, Weaver D. R. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. - PubMed
- Balsalobre A, Brown S. A, Marcacci L, Tronche F, Kellendonk C, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
- Brown S. A, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases