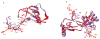Structural and functional studies of the potent anti-HIV chemokine variant P2-RANTES - PubMed (original) (raw)
Structural and functional studies of the potent anti-HIV chemokine variant P2-RANTES
Hongjun Jin et al. Proteins. 2010.
Abstract
The N-terminal region of the chemokine RANTES is critical for its function. A synthesized N-terminally modified analog of RANTES, P2-RANTES, was discovered using a phage display selection against living CCR5-expressing cells, and has been reported to inhibit HIV-1 env-mediated cell-cell fusion at subnanomolar levels (Hartley et al. J Virol 2003;77:6637-6644). In the present study we produced this protein using E. coli overexpression and extensively studied its structure and function. The x-ray crystal structure of P2-RANTES was solved and refined at 1.7 A resolution. This protein was found to be predominantly a monomer in solution by analytical ultracentrifugation, but a tetramer in the crystal. In studies of glycosaminoglycan binding, P2-RANTES was found to be significantly less able to bind heparin than wild type RANTES. We also tested this protein for receptor internalization where it was shown to be functional, in cell-cell fusion assays where recombinant P2-RANTES was a potent fusion inhibitor (IC(50) = 2.4 +/- 0.8 nM), and in single round infection assays where P2-RANTES inhibited at subnanomolar levels. Further, in a modified fusion assay designed to test specificity of inhibition, P2-RANTES was also highly effective, with a 65-fold improvement over the fusion inhibitor C37, which is closely related to the clinically approved inhibitor T-20. These studies provide detailed structural and functional information for this novel N-terminally modified chemokine mutant. This information will be very useful in the development of more potent anti-HIV agents. PDB Accession Number: 2vxw.
(c) 2009 Wiley-Liss, Inc.
Figures
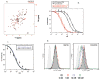
Fig. 1. Characterization and functional studies of P2-RANTES
A. The 15N HSQC of wild type RANTES (black) and P2-RANTES (red). B. The inhibition of HIV-1 fusion plotted as the percentage of fusion versus the concentration (nM) of inhibitor in the HeLa-ADA and HeLa-P5L R5 tropic cell-cell fusion assay: C37 (without 3T3 cell competition, black open circles; with 3T3, black filled circles); P2-RANTES (without 3T3 cells, red open circles; with 3T3 cells, red filled circles). Experiments were done in triplicate and repeated at least two times. Data shown is the average of all the experiments. Error bars indicate the standard deviation (± SD). Curves were fitted using KaleidaGraph. Without 3T3 cells, the 50% of fusion inhibition (IC50) for C37 and P2-RANTES are 17 ± 4.7 nM and 0.91 ± 0.5 nM respectively; With 3T3 cells, the IC50 for C37 and P2-RANTES are 61.4 ± 7.9 nM and 0.94 ± 0.5 nM respectively. The experiment is sensitive to number of cells used such that when 104 target cells were used, P2-RANTES inhibited with an IC50 of 2.4± 0.8 nM, which is also described in Results. C. The inhibition by P2-RANTES of single round virus pseudotyped with either the ADA strain (blue)or the JRFL strain (black). Data shown are the average of two independent experiments done in triplicate. Error bars indicate standard deviation. D. CCR5 internalization induced by wild type RANTES and P2-RANTES in a steady–state CCR5 down modulation FACS experiment. Data were plotted as histograms of fluorescence intensity (cell surface CCR5), with the cell number normalized for each sample. FITC fluorescence intensity from different concentrations of drug treated HeLa-TZM cells (stained with same CCR5 antibody and secondary antibody) are presented in different colors: black (0 nM); red (1 nM); blue (10 nM) and green (100 nM). The cells stained without CCR5 antibodies are showed as a gray curve.
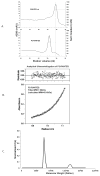
Fig. 2. GAG binding and Oligomerization state of P2-RANTES in solution
A. Heparin sepharose chromatography of wild type RANTES and P2-RANTES. Equal amounts of wild type RANTES and P2-RANTES were loaded onto a heparin sepharose column and eluted using an NaCl gradient. The dash lines indicate the NaCl concentration required for elution: 700 mM for wild type RANTES and 550mM for P2-RANTES. B. Analytical Ultracentrifugation sedimentation equilibrium experiment of P2-RANTES (24 μM protein in 50mM Bis-Tris, 200 mM NaCl, pH 5.5, at 55,000 rpm) is shown as a plot of absorbance at 280nm versus radial distance. The data is fit with a single component model with a fitted molecular weight (MW) of 7.7 kDa (Origin 3.78, Beckman). The calculated molecular weight from amino acid composition is 7.9 kDa. C. Analytical ultracentrifugation sedimentation velocity experiments of P2-RANTES were carried out at very high salt concentration to promote dimerization. This representation of the data shows the population of each species versus molecular weight. The molecular weight distribution of P2-RANTES at 50mM Bis-Tris, pH5.5, 1000 mM NaCl indicates that 90% population is in the monomer form (around 8 kDa) and 10% of the population is in the dimer form (around 16 kDa).
Fig. 3. Structure of P2-RANTES
A. Overall structure of the P2-RANTES tetramer in the crystallographic asymmetric unit. The four monomers are colored blue (A), cyan (B), green (C), and yellow (D), respectively. B. Close-up view of the N-terminal residues of monomer A. 2|Fo|−|Fc| electron density map contoured at 1.0 σ near the N-terminus of monomer A (left) showing the well-defined structure of residues from Phe0 to Ser8 (yellow stick models). On the right side, these N-terminal residues are shown in blue ribbon with white stick models. C. Close-up view of the interface between monomer A and the hydrophobic pocket of monomer C. 2|Fo|−|Fc| electron density map contoured at 1.0 σ (left) showing the interactions between the N-terminus residue Phe0 (blue stick models) of monomer A with residues Phe28, Thr30, Ala9, Val40, Cys11, and Cys50 (yellow stick models) of monomer C. On the right side, the blue Phe0 is shown interacting with monomer C (green ribbons and green stick model). D. Close-up view of the interface between monomer B and monomer C. Hydrophobic interactions between residues Tyr14(B) and Ala16(C), Ala16 (B) and Tyr14(C), are observed between monomer B (cyan ribbon) and C (green ribbons). Residue Pro2 (blue stick model) of monomer A (blue ribbon) also contacts Ile15, Val49 and Ala13 (green stick model) of monomer C. All the ribbons figures were generated using Chimera.
Fig. 4. Comparison of P2-RANTES structure to other crystal structures of RANTES
A. Overlay of the monomer structure of P2-RANTES (red) with Met-RANTES (gray, pdb file: 1eqt) and AOP-RANTES (purple, pdb file: 1b3a). The conserved four Cys residues were used to superimpose the monomer structure of these three using the “super match” function in UCSF Chimera . B. Overlay of the dimer structure of P2-RANTES with Met-RANTES and AOP-RANTES. The monomer A from P2-RANTES was used as a reference, and all dimer structures (using subunit A and B in P2-RANTES) were superimposed using UCSF Chimera . The monomer–monomer orientation in the dimer of P2-RANTES (red) is somewhat different than AOP-RANTES (purple) or Met-RANTES (gray). In particular, the changes in the N-terminal region propagate to the C-terminus, resulting in the C-terminal α-helix being oriented away from the referenced monomer. Also, Phe0 of P2-RANTES (red stick model) is pointing in a very different direction compared to the AOP group (purple stick model) in AOP-RANTES. Other residues including Ser1, Pro2 and Tyr3 (in P2-RANTES is Leu3) also show different orientations. All figures were made using UCSF Chimera .
Similar articles
- O-GalNAcylation of RANTES Improves Its Properties as a Human Immunodeficiency Virus Type 1 Entry Inhibitor.
Guan X, Chaffey PK, Chen H, Feng W, Wei X, Yang LM, Ruan Y, Wang X, Li Y, Barosh KB, Tran AH, Zhu J, Liang W, Zheng YT, Wang X, Tan Z. Guan X, et al. Biochemistry. 2018 Jan 9;57(1):136-148. doi: 10.1021/acs.biochem.7b00875. Epub 2017 Dec 14. Biochemistry. 2018. PMID: 29202246 - Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV.
Zheng Y, Han GW, Abagyan R, Wu B, Stevens RC, Cherezov V, Kufareva I, Handel TM. Zheng Y, et al. Immunity. 2017 Jun 20;46(6):1005-1017.e5. doi: 10.1016/j.immuni.2017.05.002. Immunity. 2017. PMID: 28636951 Free PMC article. - Human immunodeficiency virus type 1 entry inhibitors selected on living cells from a library of phage chemokines.
Hartley O, Dorgham K, Perez-Bercoff D, Cerini F, Heimann A, Gaertner H, Offord RE, Pancino G, Debré P, Gorochov G. Hartley O, et al. J Virol. 2003 Jun;77(12):6637-44. doi: 10.1128/jvi.77.12.6637-6644.2003. J Virol. 2003. PMID: 12767983 Free PMC article. - Highly potent chimeric inhibitors targeting two steps of HIV cell entry.
Zhao B, Mankowski MK, Snyder BA, Ptak RG, Liwang PJ. Zhao B, et al. J Biol Chem. 2011 Aug 12;286(32):28370-81. doi: 10.1074/jbc.M111.234799. Epub 2011 Jun 9. J Biol Chem. 2011. PMID: 21659523 Free PMC article. - Rational design of novel HIV-1 entry inhibitors by RANTES engineering.
Vangelista L, Secchi M, Lusso P. Vangelista L, et al. Vaccine. 2008 Jun 6;26(24):3008-15. doi: 10.1016/j.vaccine.2007.12.023. Epub 2008 Jan 10. Vaccine. 2008. PMID: 18243436 Free PMC article. Review.
Cited by
- Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Liang WG, Triandafillou CG, Huang TY, Zulueta MM, Banerjee S, Dinner AR, Hung SC, Tang WJ. Liang WG, et al. Proc Natl Acad Sci U S A. 2016 May 3;113(18):5000-5. doi: 10.1073/pnas.1523981113. Epub 2016 Apr 18. Proc Natl Acad Sci U S A. 2016. PMID: 27091995 Free PMC article. - Structural Evidence for the Tetrameric Assembly of Chemokine CCL11 and the Glycosaminoglycan Arixtra™.
Dykstra AB, Sweeney MD, Leary JA. Dykstra AB, et al. Biomolecules. 2013 Nov 6;3(4):905-22. doi: 10.3390/biom3040905. Biomolecules. 2013. PMID: 24970196 Free PMC article. - Griffithsin Retains Anti-HIV-1 Potency with Changes in gp120 Glycosylation and Complements Broadly Neutralizing Antibodies PGT121 and PGT126.
Fischer K, Nguyen K, LiWang PJ. Fischer K, et al. Antimicrob Agents Chemother. 2019 Dec 20;64(1):e01084-19. doi: 10.1128/AAC.01084-19. Print 2019 Dec 20. Antimicrob Agents Chemother. 2019. PMID: 31611356 Free PMC article. - Chemokine oligomerization in cell signaling and migration.
Wang X, Sharp JS, Handel TM, Prestegard JH. Wang X, et al. Prog Mol Biol Transl Sci. 2013;117:531-78. doi: 10.1016/B978-0-12-386931-9.00020-9. Prog Mol Biol Transl Sci. 2013. PMID: 23663982 Free PMC article. Review. - C-terminal engineering of CXCL12 and CCL5 chemokines: functional characterization by electrophysiological recordings.
Picciocchi A, Siaučiūnaiteė-Gaubard L, Petit-Hartlein I, Sadir R, Revilloud J, Caro L, Vivaudou M, Fieschi F, Moreau C, Vivès C. Picciocchi A, et al. PLoS One. 2014 Jan 31;9(1):e87394. doi: 10.1371/journal.pone.0087394. eCollection 2014. PLoS One. 2014. PMID: 24498095 Free PMC article.
References
- Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. - PubMed
- Leonard JT, Roy K. The HIV entry inhibitors revisited. Curr Med Chem. 2006;13(8):911–934. - PubMed
- Sheikine Y, Hansson GK. Chemokines and atherosclerosis. Annals of Medicine. 2004;36:98–118. - PubMed
- Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–499. - PubMed
- Gerard C, Rollins BJ. Chemokines and Disease. Nature Immunology. 2001;2:108–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical