Identification, analysis, and prediction of protein ubiquitination sites - PubMed (original) (raw)
Identification, analysis, and prediction of protein ubiquitination sites
Predrag Radivojac et al. Proteins. 2010.
Abstract
Ubiquitination plays an important role in many cellular processes and is implicated in many diseases. Experimental identification of ubiquitination sites is challenging due to rapid turnover of ubiquitinated proteins and the large size of the ubiquitin modifier. We identified 141 new ubiquitination sites using a combination of liquid chromatography, mass spectrometry, and mutant yeast strains. Investigation of the sequence biases and structural preferences around known ubiquitination sites indicated that their properties were similar to those of intrinsically disordered protein regions. Using a combined set of new and previously known ubiquitination sites, we developed a random forest predictor of ubiquitination sites, UbPred. The class-balanced accuracy of UbPred reached 72%, with the area under the ROC curve at 80%. The application of UbPred showed that high confidence Rsp5 ubiquitin ligase substrates and proteins with very short half-lives were significantly enriched in the number of predicted ubiquitination sites. Proteome-wide prediction of ubiquitination sites in Saccharomyces cerevisiae indicated that highly ubiquitinated substrates were prevalent among transcription/enzyme regulators and proteins involved in cell cycle control. In the human proteome, cytoskeletal, cell cycle, regulatory, and cancer-associated proteins display higher extent of ubiquitination than proteins from other functional categories. We show that gain and loss of predicted ubiquitination sites may likely represent a molecular mechanism behind a number of disease-associatedmutations. UbPred is available at http://www.ubpred.org.
(c) 2009 Wiley-Liss, Inc.
Figures
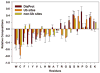
Figure 1
A Two Sample Logo of the compositional biases around Ub sites compared to the non-Ub sites. Only amino acid residues significantly enriched and depleted (P < 0.05; _t_-test) from the positive dataset are shown.
Figure 2
Relative amino acid compositions of three studied datasets. Amino acid compositions are shown relative to the composition of ordered proteins from O_PDB_S25 database. Amino acids are arranged from left to right in order of increasing flexibility as defined by Vihinen et al. The error bars represent 95% confidence intervals.
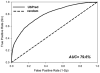
Figure 3
Receiver operating characteristic (ROC) curve for the UbPred predictor of ubiquitination sites (solid line) vs. the performance of the random model (dotted line). The area under the curve (AUC) was estimated to be 79.6%.
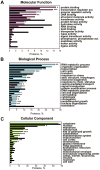
Figure 4
GO annotations for the highly ubiquitinated proteins from S. cerevisiae proteome (colored bars) with occurrence of >5% (Bonferroni corrected) as compared to the entire yeast proteome (black bars). Top 20 (whenever available) GO Slim terms are shown. A. Molecular function; B. Biological process; C. Cellular component. The proteins are arranged in order of the decreasing fraction of proteins with a specific GO annotation present in the predicted highly ubiquitinated dataset. P-values were calculated using the hypergeometric distribution and corrected for multiple hypothesis testing. *** -P<0.0001; ** - P<0.001; * - P<0.05.
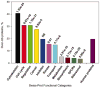
Figure 5
Frequencies of highly ubiquitinated proteins in eleven functional categories from Swiss-Prot as compared to the entire human proteome. P-values were calculated using the Wilcoxon test.
Similar articles
- A functional analysis of the yeast ubiquitin ligase Rsp5: the involvement of the ubiquitin-conjugating enzyme Ubc4 and poly-ubiquitination in ethanol-induced down-regulation of targeted proteins.
Hiraishi H, Okada M, Ohtsu I, Takagi H. Hiraishi H, et al. Biosci Biotechnol Biochem. 2009 Oct;73(10):2268-73. doi: 10.1271/bbb.90363. Epub 2009 Oct 7. Biosci Biotechnol Biochem. 2009. PMID: 19809202 - Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy.
Belgareh-Touzé N, Cavellini L, Cohen MM. Belgareh-Touzé N, et al. Autophagy. 2017 Jan 2;13(1):114-132. doi: 10.1080/15548627.2016.1252889. Epub 2016 Nov 15. Autophagy. 2017. PMID: 27846375 Free PMC article. - Identification of the endocytic sorting signal recognized by the Art1-Rsp5 ubiquitin ligase complex.
Guiney EL, Klecker T, Emr SD. Guiney EL, et al. Mol Biol Cell. 2016 Dec 15;27(25):4043-4054. doi: 10.1091/mbc.E16-08-0570. Epub 2016 Oct 19. Mol Biol Cell. 2016. PMID: 27798240 Free PMC article. - Role of Rsp5 ubiquitin ligase in biogenesis of rRNA, mRNA and tRNA in yeast.
Domanska A, Kaminska J. Domanska A, et al. RNA Biol. 2015;12(12):1265-74. doi: 10.1080/15476286.2015.1094604. RNA Biol. 2015. PMID: 26403176 Free PMC article. Review. - The role of Rsp5 ubiquitin ligase in regulation of diverse processes in yeast cells.
Kaliszewski P, Zoładek T. Kaliszewski P, et al. Acta Biochim Pol. 2008;55(4):649-62. Epub 2008 Nov 28. Acta Biochim Pol. 2008. PMID: 19039336 Review.
Cited by
- Lipopolysaccharide Primes the NALP3 Inflammasome by Inhibiting Its Ubiquitination and Degradation Mediated by the SCFFBXL2 E3 Ligase.
Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, Gibson KF, Zou C, Chen BB, Mallampalli RK. Han S, et al. J Biol Chem. 2015 Jul 17;290(29):18124-18133. doi: 10.1074/jbc.M115.645549. Epub 2015 Jun 2. J Biol Chem. 2015. PMID: 26037928 Free PMC article. - Hrq1/RECQL4 regulation is critical for preventing aberrant recombination during DNA intrastrand crosslink repair and is upregulated in breast cancer.
Luong TT, Li Z, Priedigkeit N, Parker PS, Böhm S, Rapchak K, Lee AV, Bernstein KA. Luong TT, et al. PLoS Genet. 2022 Sep 20;18(9):e1010122. doi: 10.1371/journal.pgen.1010122. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36126066 Free PMC article. - Individual Src-family tyrosine kinases direct the degradation or protection of the clock protein Timeless via differential ubiquitylation.
O'Reilly LP, Zhang X, Smithgall TE. O'Reilly LP, et al. Cell Signal. 2013 Apr;25(4):860-6. doi: 10.1016/j.cellsig.2012.12.009. Epub 2012 Dec 22. Cell Signal. 2013. PMID: 23266470 Free PMC article. - Precise Prediction of Calpain Cleavage Sites and Their Aberrance Caused by Mutations in Cancer.
Liu ZX, Yu K, Dong J, Zhao L, Liu Z, Zhang Q, Li S, Du Y, Cheng H. Liu ZX, et al. Front Genet. 2019 Aug 8;10:715. doi: 10.3389/fgene.2019.00715. eCollection 2019. Front Genet. 2019. PMID: 31440276 Free PMC article. - Regulation of Blood Pressure by Targeting CaV1.2-Galectin-1 Protein Interaction.
Hu Z, Li G, Wang JW, Chong SY, Yu D, Wang X, Soon JL, Liang MC, Wong YP, Huang N, Colecraft HM, Liao P, Soong TW. Hu Z, et al. Circulation. 2018 Oct 2;138(14):1431-1445. doi: 10.1161/CIRCULATIONAHA.117.031231. Circulation. 2018. PMID: 29650545 Free PMC article.
References
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195–201. - PubMed
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8(3):499–504. - PubMed
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4(3):192–201. - PubMed
- Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002;12(12):569–579. - PubMed
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1(2):193–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases