MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation - PubMed (original) (raw)
MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation
Guisheng Song et al. J Biol Chem. 2009.
Abstract
MicroRNAs contribute to cancer development by acting as oncogenes or tumor suppressor genes. However, only a few microRNA target genes were determined. We identified a nearly perfect complementarity between miR-206 and the 3'-untranslated regions of both mouse and human notch3. Expression of miR-206 decreased the luciferase activity dose-dependently when cotransfected with the mouse or human notch3 3'-untranslated region-luciferase reporter containing the miR-206 target site in HeLa cells. This suppression was relieved by deletion and mutation of the miR-206-binding site and was partially recovered by expression of notch3 or by a specific inhibitor of miR-206. Interestingly, overexpression of miR-206 decreased the levels of both Notch3 protein and mRNA. Expression of miR-206 markedly induced apoptotic cell death and blocked the anti-apoptotic activity of Notch3. In addition, ectopic expression of miR-206 inhibited HeLa cell migration and focus formation. Therefore, we identified miR-206 as a pro-apoptotic activator of cell death, which was associated with its inhibition of notch3 signaling and tumor formation. The inhibition of cancer cell migration and focus formation by miR-206 strongly suggests that miR-206 may function as a novel tumor suppressor.
Figures
FIGURE 1.
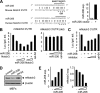
miR-206 targets and regulates mouse notch3. A: left, sequence alignment between miR-206 and the 3′-UTR of notch3 in mouse and human. Solid line, seed match region; dashed line, seed-deleted region. Right, real-time PCR analysis of miR-206 expression (exp.) in HeLa cells transfected with a control (−) or miR-206 (+; 400 ng) expression vector. B: HeLa cells cotransfected with a luciferase reporter containing the mouse notch3 3′-UTR (left) or the miR-206-binding site-deleted (del) mouse notch3 3′-UTR (right), a miR-206 expression plasmid (50, 100, and 200 ng), or a full-length notch3 expression plasmid (50 and 100 ng). cel-miR-64 (200 ng) is a non-related miRNA that did not target notch3 and was used as a negative control. Luciferase activities were measured 24 h post-transfection and normalized to the corresponding vector control. Each transfection was done in triplicate, and the experiment was repeated three times. A representative experiment is shown (mean ± S.E.). Luc./gal. activity, luciferase/galactosidase activity. C: luciferase activities determined in HeLa cells transfected with a mouse notch3 3′-UTR-reporter in the presence or absence of the miR-206 expression plasmid (200 ng) or a miR-206 inhibitor (final concentrations of 10, 20, and 40 pmol). Data are represented as the mean ± S.E. D: immunoblot showing that miR-206 (4 μg, 6-cm plate) induced a decrease in endogenous mouse Notch3 protein in mouse MEFs. β-Actin was used as a loading control. cel-miR-64 (4 μg), a non-related miRNA that did not target notch3, was used as a negative control. E: real-time PCR analysis of miR-206 and mouse notch3 expression in SHP+/+ MEFs overexpressed with a miR-206 expression plasmid (50, 100, and 200 ng). Data are represented as the mean ± S.E.
FIGURE 2.
miR-206 targets and regulates human notch3. A: HeLa cells cotransfected with a luciferase reporter construct containing the human notch3 3′-UTR (left) or the miR-206 target site-deleted (del) human notch3 3′-UTR (right), a miR-206 expression plasmid (50, 100, and 200 ng), or a full-length notch3 expression plasmid (50 and 100 ng), and luciferase activities were determined. cel-miR-64 (200 ng) was a non-related miRNA that did not target human notch3 and was used as a negative control. Luc./gal. activity, luciferase/galactosidase activity. B: left, luciferase activities determined in HeLa cells transfected with a human notch3 3′-UTR-reporter in the presence or absence of a miR-206 expression plasmid (200 ng) and a miR-206 inhibitor (final concentrations of 10, 20, and 40 pmol). Right, real-time PCR analysis of miR-206 expression (exp.) in HeLa cells transfected with a control vector (−), miR-206 expression vector (100 ng), or miR-206 inhibitor (final concentration of 40 pmol). C: immunoblot showing that miR-206 (4 μg, 6-cm plate) induced a decrease in endogenous human Notch3 protein in human HeLa cells. β-Actin was used as a loading control. D: real-time PCR analysis of miR-206 and human notch3 expression in HeLa cells overexpressed with a miR-206 expression plasmid (3 and 6 μg, 6-cm plate). E: luciferase activities determined in SHP+/+ (wild-type) and _SHP_−/− (SHP knock-out) MEFs transfected with the mouse or human notch3 3′-UTR or glucokinase (GK) 3′-UTR. All data are represented as the mean ± S.E.
FIGURE 3.
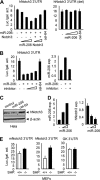
miR-206 activates apoptosis. A: left, real-time PCR analysis of miR-206 expression (exp.). Middle, annexin V staining and flow cytometry analysis of cell death. The lower right quadrant of each plot contains early apoptotic cells. This experiment was repeated three independent times, and similar results were obtained each time. Right, caspase 3 (cas. 3) activity assays. 7-AAD, 7-aminoactinomycin; PE, phycoerythrin. B: left, annexin V staining and flow cytometry analysis of cell death. Middle, real-time PCR analysis of miR-206 expression. Right, semiquantitative PCR analysis of notch3 mRNA expression. All experiments were performed in HeLa cells (1 × 106, 6-cm plate) transfected with a miR-206 expression vector (6 μg) in the absence or presence of its inhibitor (final concentration of 40 pmol) or with a full-length notch3 (N) expression vector (6 μg) alone or in combination with miR-206 (M). C, control.
FIGURE 4.
miR-206 inhibits tumor cell migration. A wound healing assay was performed on HeLa cells (1 × 106, 6-cm plate) transfected with either an empty vector (control) or a miR-206 expression vector (6 μg) in the absence or presence of the miR-206 inhibitor (final concentration of 40 pmol) or with a full-length notch3 expression vector (6 μg) alone or in combination with miR-206. The experiments were repeated three times with similar results, and one representative experiment is shown. Wound closure was photographed and quantified as shown. The residual gap between the migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area.
FIGURE 5.
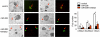
miR-206 inhibits focus formation. Left, focus formation assay performed on HeLa cells (0.5 × 106, 35-mm plate) transfected with either a GFP-control vector or a GFP-miR-206 expression vector (3 μg) in the absence or presence of the miR-206 inhibitor (final concentration of 40 pmol). GFP signal indicates foci formed from cells transfected with the GFP-control or GFP-miR-206 vector. Right, statistical analysis of foci formed. Data are represented as the mean ± S.E. (n = three fields/plate of three plates). *, p < 0.01.
Similar articles
- MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer.
Wang XW, Xi XQ, Wu J, Wan YY, Hui HX, Cao XF. Wang XW, et al. Oncol Rep. 2015 Mar;33(3):1402-10. doi: 10.3892/or.2015.3731. Epub 2015 Jan 19. Oncol Rep. 2015. PMID: 25607234 - The miR-1-NOTCH3-Asef pathway is important for colorectal tumor cell migration.
Furukawa S, Kawasaki Y, Miyamoto M, Hiyoshi M, Kitayama J, Akiyama T. Furukawa S, et al. PLoS One. 2013 Nov 11;8(11):e80609. doi: 10.1371/journal.pone.0080609. eCollection 2013. PLoS One. 2013. PMID: 24244701 Free PMC article. - MicroRNA-7a regulates Müller glia differentiation by attenuating Notch3 expression.
Baba Y, Aihara Y, Watanabe S. Baba Y, et al. Exp Eye Res. 2015 Sep;138:59-65. doi: 10.1016/j.exer.2015.06.022. Epub 2015 Jun 26. Exp Eye Res. 2015. PMID: 26122050 - Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150.
Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, Mussolin L, Ruggero K, Bonanno L, Guffanti A, De Bellis G, Gerosa G, Stellin G, D'Agostino DM, Basso G, Bronte V, Indraccolo S, Amadori A, Zanovello P. Ghisi M, et al. Blood. 2011 Jun 30;117(26):7053-62. doi: 10.1182/blood-2010-12-326629. Epub 2011 May 6. Blood. 2011. PMID: 21551231 - Suppression of p53 by Notch3 is mediated by Cyclin G1 and sustained by MDM2 and miR-221 axis in hepatocellular carcinoma.
Giovannini C, Minguzzi M, Baglioni M, Fornari F, Giannone F, Ravaioli M, Cescon M, Chieco P, Bolondi L, Gramantieri L. Giovannini C, et al. Oncotarget. 2014 Nov 15;5(21):10607-20. doi: 10.18632/oncotarget.2523. Oncotarget. 2014. PMID: 25431954 Free PMC article.
Cited by
- Role of microRNAs in breast cancer.
Singh R, Mo YY. Singh R, et al. Cancer Biol Ther. 2013 Mar;14(3):201-12. doi: 10.4161/cbt.23296. Epub 2013 Jan 4. Cancer Biol Ther. 2013. PMID: 23291983 Free PMC article. Review. - MicroRNAs in cancer treatment and prognosis.
Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. Schoof CR, et al. Am J Cancer Res. 2012;2(4):414-33. Epub 2012 Jun 28. Am J Cancer Res. 2012. PMID: 22860232 Free PMC article. - miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells.
Macquarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ. Macquarrie KL, et al. Skelet Muscle. 2012 Apr 29;2(1):7. doi: 10.1186/2044-5040-2-7. Skelet Muscle. 2012. PMID: 22541669 Free PMC article. - MicroRNA-710 regulates multiple pathways of carcinogenesis in murine metastatic breast cancer.
Yoo B, Meka N, Sheedy P, Billig AM, Pantazopoulos P, Medarova Z. Yoo B, et al. PLoS One. 2019 Dec 13;14(12):e0226356. doi: 10.1371/journal.pone.0226356. eCollection 2019. PLoS One. 2019. PMID: 31834924 Free PMC article. - Puerarin promotes the proliferation and differentiation of MC3T3-E1 cells via microRNA-106b by targeting receptor activator of nuclear factor-κB ligand.
Shan Z, Cheng N, Huang R, Zhao B, Zhou Y. Shan Z, et al. Exp Ther Med. 2018 Jan;15(1):55-60. doi: 10.3892/etm.2017.5405. Epub 2017 Oct 31. Exp Ther Med. 2018. PMID: 29375675 Free PMC article.
References
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 - PubMed
- Bartel D. P. (2004) Cell 116, 281–297 - PubMed
- Du T., Zamore P. D. (2005) Development 132, 4645–4652 - PubMed
- Wienholds E., Plasterk R. H. (2005) FEBS Lett. 579, 5911–5922 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous