Protein S-nitrosylation in health and disease: a current perspective - PubMed (original) (raw)
Review
Protein S-nitrosylation in health and disease: a current perspective
Matthew W Foster et al. Trends Mol Med. 2009 Sep.
Abstract
Protein S-nitrosylation constitutes a large part of the ubiquitous influence of nitric oxide on cellular signal transduction and accumulating evidence indicates important roles for S-nitrosylation both in normal physiology and in a broad spectrum of human diseases. Here we review recent findings that implicate S-nitrosylation in cardiovascular, pulmonary, musculoskeletal and neurological (dys)function, as well as in cancer. The emerging picture shows that, in many cases, pathophysiology correlates with hypo- or hyper-S-nitrosylation of specific protein targets rather than a general cellular insult due to loss of or enhanced nitric oxide synthase activity. In addition, it is increasingly evident that dysregulated S-nitrosylation can not only result from alterations in the expression, compartmentalization and/or activity of nitric oxide synthases, but can also reflect a contribution from denitrosylases, including prominently the S-nitrosoglutathione (GSNO)-metabolizing enzyme GSNO reductase. Finally, because exogenous mediators of protein S-nitrosylation or denitrosylation can substantially affect the development or progression of disease, potential therapeutic agents that modulate S-nitrosylation could well have broad clinical utility.
Figures
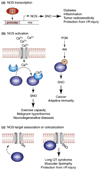
Fig. 1. NOS-dependent mechanisms of _S_-nitrosylation
Three principal mechanisms for regulation of NO synthase (NOS)-dependent protein _S_-nitrosylation and the (patho)physiology associated with the induction or disruption of these mechanisms are shown. (a) Binding of a transcription factor (TF) to NOS promoter induces expression of the gene encoding NOS. (b) (Left) influx into the cytosol of extracellular or internal store-derived Ca2+ promotes Ca2+-calmodulin (CaM) binding and activation of eNOS and nNOS. Alternatively (right), phosphoinositide-3 kinase (PI3K) activates protein kinase B (Akt), which phosphorylates and activates eNOS. (c) Subcellular compartmentation (co-localization) of NOS and its substrates, which may involve a direct interaction (as in the illustrated case of a membrane-intercalated ion channel) is an important determinant of the target specificity of _S_-nitrosylation, and dysregulated co-localization can result in hyper- or hypo-_S_-nitrosylation.
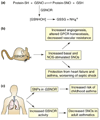
Fig. 2. Regulation of SNO homeostasis by _S_-nitrosoglutathione reductase (GSNOR)
(a) In cells, the low-mass _S_-nitrosothiol, _S_-nitrosoglutathione (GSNO), is in equilibrium with a subset of protein _S_-nitrosothiols. GSNO is metabolized by the enzyme GSNOR, and cells and tissues lacking GSNOR exhibit increased levels of SNO-proteins. (b) Analyses in GSNOR-knockout mice (GSNOR−/−) have revealed numerous roles for protein _S_-nitrosylation. In particular, knockout animals exhibit low systemic vascular resistance and increased cardiac output under basal conditions, and are protected from mocardial infarction and allergen-induced airway hyperreactivity. (c) Mutations in GSNOR, as well as increased airway GSNOR expression and activity, are associated with human asthma.
Similar articles
- Protein S-Nitrosylation: Determinants of Specificity and Enzymatic Regulation of S-Nitrosothiol-Based Signaling.
Stomberski CT, Hess DT, Stamler JS. Stomberski CT, et al. Antioxid Redox Signal. 2019 Apr 1;30(10):1331-1351. doi: 10.1089/ars.2017.7403. Epub 2018 Jan 10. Antioxid Redox Signal. 2019. PMID: 29130312 Free PMC article. Review. - Regulation of DNA repair by S-nitrosylation.
Tang CH, Wei W, Liu L. Tang CH, et al. Biochim Biophys Acta. 2012 Jun;1820(6):730-5. doi: 10.1016/j.bbagen.2011.04.014. Epub 2011 May 5. Biochim Biophys Acta. 2012. PMID: 21571039 Free PMC article. Review. - Protein denitrosylation: enzymatic mechanisms and cellular functions.
Benhar M, Forrester MT, Stamler JS. Benhar M, et al. Nat Rev Mol Cell Biol. 2009 Oct;10(10):721-32. doi: 10.1038/nrm2764. Epub 2009 Sep 9. Nat Rev Mol Cell Biol. 2009. PMID: 19738628 Review. - Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease.
Anand P, Stamler JS. Anand P, et al. J Mol Med (Berl). 2012 Mar;90(3):233-44. doi: 10.1007/s00109-012-0878-z. Epub 2012 Feb 24. J Mol Med (Berl). 2012. PMID: 22361849 Free PMC article. Review. - Nebivolol Acts as a S-Nitrosoglutathione Reductase Inhibitor: A New Mechanism of Action.
Jiang H, Polhemus DJ, Islam KN, Torregrossa AC, Li Z, Potts A, Lefer DJ, Bryan NS. Jiang H, et al. J Cardiovasc Pharmacol Ther. 2016 Sep;21(5):478-85. doi: 10.1177/1074248415626300. Epub 2016 Jan 8. J Cardiovasc Pharmacol Ther. 2016. PMID: 26746429
Cited by
- Thiol-redox signaling, dopaminergic cell death, and Parkinson's disease.
Garcia-Garcia A, Zavala-Flores L, Rodriguez-Rocha H, Franco R. Garcia-Garcia A, et al. Antioxid Redox Signal. 2012 Dec 15;17(12):1764-84. doi: 10.1089/ars.2011.4501. Epub 2012 May 3. Antioxid Redox Signal. 2012. PMID: 22369136 Free PMC article. Review. - S-Nitrosation Mediates Multiple Pathways That Lead to Tumor Progression in Estrogen Receptor-Negative Breast Cancer.
Switzer CH, Ridnour LA, Cheng R, Heinecke J, Burke A, Glynn S, Ambs S, Wink DA. Switzer CH, et al. For Immunopathol Dis Therap. 2012;3(2):117-124. doi: 10.1615/ForumImmunDisTher.2012006108. For Immunopathol Dis Therap. 2012. PMID: 23543871 Free PMC article. - S-nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer.
Switzer CH, Glynn SA, Cheng RY, Ridnour LA, Green JE, Ambs S, Wink DA. Switzer CH, et al. Mol Cancer Res. 2012 Sep;10(9):1203-15. doi: 10.1158/1541-7786.MCR-12-0124. Epub 2012 Aug 9. Mol Cancer Res. 2012. PMID: 22878588 Free PMC article. - Protein Oxidative Modifications: Beneficial Roles in Disease and Health.
Cai Z, Yan LJ. Cai Z, et al. J Biochem Pharmacol Res. 2013 Mar;1(1):15-26. J Biochem Pharmacol Res. 2013. PMID: 23662248 Free PMC article. - H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response.
Krishnan N, Fu C, Pappin DJ, Tonks NK. Krishnan N, et al. Sci Signal. 2011 Dec 13;4(203):ra86. doi: 10.1126/scisignal.2002329. Sci Signal. 2011. PMID: 22169477 Free PMC article.
References
- Hess DT, et al. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. - PubMed
- Foster MW, et al. S-nitrosylation in health and disease. Trends Mol. Med. 2003;9:160–168. - PubMed
- Jaffrey SR, et al. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. - PubMed
- Gow AJ, et al. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 2002;277:9637–9640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL091876/HL/NHLBI NIH HHS/United States
- HL059130/HL/NHLBI NIH HHS/United States
- R01 HL091876-01A1/HL/NHLBI NIH HHS/United States
- U19 ES012496/ES/NIEHS NIH HHS/United States
- U19 ES012496-05/ES/NIEHS NIH HHS/United States
- U19-ES012496/ES/NIEHS NIH HHS/United States
- R01 HL066179-01/HL/NHLBI NIH HHS/United States
- HL075443/HL/NHLBI NIH HHS/United States
- R01 HL059130-11/HL/NHLBI NIH HHS/United States
- R01 HL059130/HL/NHLBI NIH HHS/United States
- P01 HL075443/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources