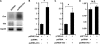Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation - PubMed (original) (raw)
Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation
Na Lian et al. J Biol Chem. 2009.
Abstract
Activating transcription factor 4 (ATF4) is an osteoblast-enriched transcription factor that regulates osteocalcin transcription and osteoblast terminal differentiation. To identify functional partners of ATF4, we applied ROS17/2.8 osteoblast nuclear extracts and purified recombinant His-ATF4 onto a Ni(+) affinity matrix chromatography column. Vimentin was identified by liquid chromatography-mass spectrometry. Coimmunoprecipitation and pulldown assays revealed that vimentin interacted with ATF4 with its first leucine zipper domain. DNA cotransfection and gel retardation demonstrated that vimentin inhibited the transactivation activity of ATF4 on osteocalcin by preventing it to bind OSE1, the ATF4 binding site on the osteocalcin promoter. Northern hybridization revealed that vimentin was expressed at a high level in immature osteoblasts and a low level in fully differentiated osteoblasts. Down-regulation of vimentin by small interfering RNA induced endogenous osteocalcin transcription in immature osteoblasts. Conversely, ectopic overexpression of vimentin in osteoblasts inhibited osteoblast differentiation as shown by lower alkaline phosphatase activity, delayed mineralization, and decreased expression of osteoblast marker genes such as bone sialoprotein and osteocalcin. Together, our data uncover a novel mechanism whereby a cytoskeletal protein, vimentin, acts as a break on differentiation in immature osteoblasts by interacting with ATF4.
Figures
FIGURE 1.
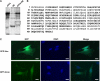
Identification of vimentin. A, pulldown assay using His-ATF4 (ATF4, lane 1) as bait and NEs from ROS17/2.8 osteoblastic cells (lane 2) as pool of proteins. Proteins were separated with a 4–20% gradient SDS-PAGE and stained with Coomassie Brilliant Blue. A 37 kDa band (boxed) was excised and analyzed by MS. B, amino acid sequence of vimentin. MS analysis revealed peptides (bold letters) that covered 54% of vimentin. C, GFP-tagged vimentin (upper panel) transfected into MC3T3-E1 osteoblastic cells and analyzed for subcellular localization in comparison with GFP vector (lower panel) alone. Nuclei were visualized using DAPI staining (middle panel). Scale bar, 10 μm.
FIGURE 2.
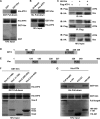
Vimentin (Vim) interacts with ATF4. A, pulldown assay showing that GST-vimentin binds directly to ATF4. B, pulldown assay using His-ATF4 and Ni-NTA resin column showing that ATF4 binds directly to vimentin. C, reciprocal coimmunoprecipitation. NEs of COS cells transfected with HA-vimentin and/or FLAG-ATF4 were immunoprecipitated with anti-HA or anti-FLAG antibody and visualized with anti-FLAG and anti-HA antibodies (top two panels). Total protein (10%) of each transfection served as loading control (bottom panel). D, schematic illustration of ATF4 primary structure showing two LZ domains at N and C termini. Letter b represents basic amino acid-rich DNA binding domain. E, schematic presentation of vimentin primary structure showing three PLZs of vimentin. F, PLZ1 of vimentin interacting with ATF4. Pulldown assay was carried out using purified GST-vimentin and its truncated variants GST-Vim1–3 and His-ATF4. Note that only full-length vimentin (Vim-fl) or its variant containing the first PLZ interacted with His-ATF4. G, LZs of ATF4 interacting with vimentin. Pulldown assay was performed using purified His-ATF4 and its three indicated deletional variants. Note that full-length ATF4 (Full length) and both of its LZ-containing variants, 1–151 and 186–349, bound vimentin.
FIGURE 3.
Vimentin (Vim) inhibits ATF4-dependent transactivation of osteocalcin transcription by blocking its binding to OSE1. A–C, DNA cotransfections of COS1 cells with reporter construct containing six copies of ATF4 binding site OSE1, p6OSE1-Luc (A), a native osteocalcin promoter, p160-Luc (B), three copies of binding sites of AP1 family (C), and/or expression vectors of ATF4, FosB, and/or vimentin as indicated. Note that ATF4-induced but not FosB-induced luciferase activity was inhibited by more than 70% by vimentin. D, vimentin inhibiting the binding of ATF4 to OSE1. EMSA was performed using purified His-ATF4 and radiolabeled OSE1 as a probe. Note that vimentin inhibited ATF4 binding to OSE1 dose-dependently. E, EMSA showing that vimentin inhibits endogenous ATF4 (middle bands are ATF4·OSE1 complexes (9)) from ROS17/2.8 osteoblastic cells binding to OSE1. F, DNA cotransfection in COS1 cells with p6OSE1-Luc and expression vectors of FLAG-ATF4 and/or HA-Vim1, -Vim2, or -Vim3. G and H, EMSA. Note that GST-Vim1 inhibited His-ATF4 (G) or endogenous ATF4 (H) binding to OSE1 in a dose-dependent manner. The luciferase activities were normalized for the β-galactosidase activity and are presented as fold activation relative to the luciferase levels of the reporter construct alone. Values are the mean ± S.D. of three independent experiments. *, p < 0.01; **, p < 0.001; N.S., not significant.
FIGURE 4.
Down-regulation of vimentin by siRNA enhances osteocalcin expression. A–C, DNA cotransfections of 2T3 osteoblastic cells with reporter p6OSE1-Luc (A), p160-Luc (B), AP1-Luc (C), and vectors of control (psiRNA) or vimentin (psiRNA-Vim). Note that vimentin siRNA induced activation of osteocalcin promoter by >2-fold. Values are the mean ± S.D. of three independent experiments. *, p < 0.01; N.S., not significant. D, Northern blot using total RNA from 2T3 osteoblasts transfected with control or siRNA against vimentin. Note that depletion of vimentin triggered osteocalcin expression. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was used as a loading control.
FIGURE 5.
Vimentin delays osteoblast differentiation. A, overexpression of vimentin in MC3T3-E1 osteoblasts. Vimentin in pcDNA3.1 expression vector was stably expressed in the MC3T3-E1 osteoblastic cell line. Bovine growth hormone (BGH) was used to probe for exogenous vimentin. B, vimentin decreasing alkaline phosphatase (ALP) activity. Decreased alkaline phosphatase activity at 2 and 4 days after osteogenic induction was observed in MC3T3-E1 cells overexpressing vimentin compared with the controls. C, vimentin inhibiting mineralization of MC3T3-E1 cells. von Kossa staining was done at 6, 12, and 20 days after differentiation of control cells (Vector) and vimentin-overexpressing cells. Cells were counterstained with acid fuchsin on day 0 before osteogenic differentiation induction showing that the same amount of cells were plated. D, Northern blot analysis showing the expression of osteocalcin (Ocn) and bone sialoprotein (Bsp), two mature osteoblast marker genes, and Col1_α_1, an early osteoblast marker, together with three transcription factors that are important for osteoblast differentiation, Runx2, osterix (Osx), and ATF4 (Atf4). Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was used as a loading control.
FIGURE 6.
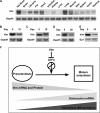
Expression pattern of vimentin (Vim) mRNA and protein. A, Northern blot analysis using total RNA from indicated mouse tissues showing that vimentin mRNA is highly expressed in bone. B–D. Northern blot using total RNA from MC3T3-E1 (B), primary calvarial osteoblasts (C), and primary bone marrow stromal cells (D) at the indicated times after differentiation induction. Note that vimentin mRNA is down-regulated at day 10 or day 20 after osteoblast differentiation induction. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was used as a loading control. E, Western blot analysis using nuclear extracts of rat primary calvarial osteoblasts. Note that vimentin protein decreased during osteoblast differentiation. These results are representatives of three independent experiments. F, schematic illustration of the role of vimentin in osteoblast differentiation. In preosteoblasts, vimentin is highly expressed, and it inhibits osteocalcin transcription and premature osteoblast differentiation by binding to ATF4. Along with the progression of osteoblast differentiation, the vimentin level decreases, and thus the inhibition of ATF4 is released, osteocalcin transcription is then initiated, and osteoblast terminal differentiation is achieved.
Similar articles
- Transforming growth factor β suppresses osteoblast differentiation via the vimentin activating transcription factor 4 (ATF4) axis.
Lian N, Lin T, Liu W, Wang W, Li L, Sun S, Nyman JS, Yang X. Lian N, et al. J Biol Chem. 2012 Oct 19;287(43):35975-84. doi: 10.1074/jbc.M112.372458. Epub 2012 Sep 4. J Biol Chem. 2012. PMID: 22952236 Free PMC article. - Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression.
Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Xiao G, et al. J Biol Chem. 2005 Sep 2;280(35):30689-96. doi: 10.1074/jbc.M500750200. Epub 2005 Jul 5. J Biol Chem. 2005. PMID: 16000305 - CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4.
Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. Tominaga H, et al. Mol Biol Cell. 2008 Dec;19(12):5373-86. doi: 10.1091/mbc.e08-03-0329. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843047 Free PMC article. - Identification of additional dimerization partners of FIAT, the factor inhibiting ATF4-mediated transcription.
St-Arnaud R, Elchaarani B. St-Arnaud R, et al. Ann N Y Acad Sci. 2007 Nov;1116:208-15. doi: 10.1196/annals.1402.028. Ann N Y Acad Sci. 2007. PMID: 18083929 Review. - FIAT control of osteoblast activity.
St-Arnaud R, Mandic V. St-Arnaud R, et al. J Cell Biochem. 2010 Feb 15;109(3):453-9. doi: 10.1002/jcb.22435. J Cell Biochem. 2010. PMID: 20013796 Review.
Cited by
- Transforming growth factor β suppresses osteoblast differentiation via the vimentin activating transcription factor 4 (ATF4) axis.
Lian N, Lin T, Liu W, Wang W, Li L, Sun S, Nyman JS, Yang X. Lian N, et al. J Biol Chem. 2012 Oct 19;287(43):35975-84. doi: 10.1074/jbc.M112.372458. Epub 2012 Sep 4. J Biol Chem. 2012. PMID: 22952236 Free PMC article. - Combinatorial control of ATF4-dependent gene transcription in osteoblasts.
St-Arnaud R, Hekmatnejad B. St-Arnaud R, et al. Ann N Y Acad Sci. 2011 Nov;1237:11-8. doi: 10.1111/j.1749-6632.2011.06197.x. Ann N Y Acad Sci. 2011. PMID: 22082360 Free PMC article. Review. - Vimentin in cancer and its potential as a molecular target for cancer therapy.
Satelli A, Li S. Satelli A, et al. Cell Mol Life Sci. 2011 Sep;68(18):3033-46. doi: 10.1007/s00018-011-0735-1. Epub 2011 Jun 3. Cell Mol Life Sci. 2011. PMID: 21637948 Free PMC article. Review. - Structure and function of vimentin in the generation and secretion of extracellular vimentin in response to inflammation.
Yuan Z, Janmey PA, McCulloch CA. Yuan Z, et al. Cell Commun Signal. 2025 Apr 18;23(1):187. doi: 10.1186/s12964-025-02194-z. Cell Commun Signal. 2025. PMID: 40251523 Free PMC article. Review. - Aberrant MEK5/ERK5 signalling contributes to human colon cancer progression via NF-κB activation.
Simões AE, Pereira DM, Gomes SE, Brito H, Carvalho T, French A, Castro RE, Steer CJ, Thibodeau SN, Rodrigues CM, Borralho PM. Simões AE, et al. Cell Death Dis. 2015 Apr 9;6(4):e1718. doi: 10.1038/cddis.2015.83. Cell Death Dis. 2015. PMID: 25855966 Free PMC article.
References
- Banerjee C., McCabe L. R., Choi J. Y., Hiebert S. W., Stein J. L., Stein G. S., Lian J. B. (1997) J. Cell. Biochem. 66, 1–8 - PubMed
- Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 - PubMed
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 - PubMed
- Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 - PubMed
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) Cell 108, 17–29 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials