Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics - PubMed (original) (raw)
Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics
Ignazio Cali et al. Brain. 2009 Oct.
Abstract
Five phenotypically distinct subtypes have been identified in sporadic Creutzfeldt-Jakob disease (sCJD), based on the methionine/valine polymorphic genotype of codon 129 of the prion protein (PrP) gene and the presence of either one of the two protease K-resistant scrapie prion protein (PrP(Sc)) types identified as 1 and 2. The infrequent co-existence of both PrP(Sc) types in the same case has been known for a long time. Recently, it has been reported, using type-specific antibodies, that the PrP(Sc) type 1 is present in all cases of sCJD carrying PrP(Sc) type 2. The consistent co-occurrence of both PrP(Sc) types complicates the diagnosis and the current classification of sCJD, and has implications for the pathogenesis of naturally occurring prion diseases. In the present study, we investigated the prevalence of PrP(Sc) types 1 and 2 co-occurrence, along with its effects on the disease phenotype and PrP(Sc) strain characteristics, comparatively analysing 34 cases of sCJD, all methionine homozygous at codon 129 of the PrP gene (sCJDMM). To minimize overestimating the prevalence of the sCJDMM cases carrying PrP(Sc) types 1 and 2 (sCJDMM1-2), we used proteinase K concentrations designed to hydrolyse all fragments resulting from an incomplete digestion, while preserving the protease-resistant PrP(Sc) core. Furthermore, we used several antibodies to maximize the detection of both PrP(Sc) types. Our data show that sCJDMM cases associated exclusively with either PrP(Sc) type 1 (sCJDMM1) or PrP(Sc) type 2 (sCJDMM2) do exist; we estimate that they account for approximately 56% and 5% of all the sCJDMM cases, respectively; while in 39% of the cases, both PrP(Sc) types 1 and 2 are present together (sCJDMM1-2) either mixed in the same anatomical region or separate in different regions. Clinically, sCJDMM1-2 had an average disease duration intermediate between the other two sCJDMM subtypes. The histopathology was also intermediate, except for the cerebellum where it resembled that of sCJDMM1. These features, along with the PrP immunostaining pattern, offer a diagnostic clue. We also observed a correlation between the disease duration and the prevalence of PrP(Sc) type 2 and sCJDMM2 phenotypes. The use of different antibodies and of the conformational stability immunoassay indicated that the co-existence of types 1 and 2 in the same anatomical region may confer special conformational characteristics to PrP(Sc) types 1 and 2. All of these findings indicate that sCJDMM1-2 should be considered as a separate entity at this time.
Figures
Figure 1
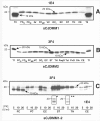
Validation of PrPSc typing by protease digestion in sCJDMM2 and sCJDMM1. S1 fractions from the frontal cortex were incubated with several amounts of PK and probed with 3F4 (A, C) or 12B2 (B). (A) The core of the unglycosylated PrPSc type 2 (large black solid arrow) is detectable after treatment with up to 40 U/ml PK (up to 160 U/ml in long exposure films). The partially digested PrP fragments (dashed arrows) are visible between 0.15 (denoted by asterisk) and 0.6 U/ml of PK treatment but are completely hydrolyzed at relatively low PK concentrations ranging from 2.5 to 5 U/ml (denoted by double asterisk). The bands indicated by the black solid arrows in A* presumably represent monoglycosylated PK-resistant PrPSc fragments. (B) The mAb 12B2, which binds to the PrPSc type 1 but not to type 2 PK-resistant fragments, immunodetects the four partially digested PrP fragments but not the unglycosylated PrPSc type 2 core confirming that the fragments are incompletely digested, while the ‘core’ belongs to type 2. (C) Four partially cleaved PrP fragments (dashed arrows) are detectable between 0.15 and 0.6 U/ml PK in sCJDMM1 preparations. The core of the unglycosylated PrPSc type 1 (solid grey arrow) is resistant up to 40 U/ml (or up to 80 U/ml PK in long exposure films). For the small black solid arrows see (A*).
Figure 2
Diagram of epitopes of various anti-PrP antibodies and western blotting of PrPSc mixed naturally and artificially. (A) Representation of PK cleavage sites, PK-sensitive and PK-resistant regions and epitope location in human PrPSc. Long and short arrows identify the primary and secondary PK cleavage sites located along the variable region G74-S103. This region is depicted out of proportion. The locations of the relevant residues for PrPSc types 1 and 2 are indicated with white and black arrows, respectively. Tryptophan 89 (W89*) represents the expected PK cleavage site (see PeptideCutter Program) generating the PrPSc type 1 fragment of ∼20 kDa when samples are homogenized at pH ∼8.0. The curly brackets indicate the PrP epitopes recognized by the antibodies used in this study. The basis for the reactivity of mAb 12B2 with PrPSc type 1 but not type 2 is illustrated with the dotted lines. (B) Both mAb 3F4 and 1E4 immunoreacted with PrPSc type 2 that had been artificially mixed with PrPSc type 1 (lanes 1 and 4). In contrast, PrPSc type 2 from sCJDMM1-2, where types 1 and 2 co-occurred in the same brain region, was detected by 1E4 only (lanes 5 and 6). Lanes 1 and 4 were generated with tissue from the frontal cortex of sCJDMM1 and sCJDMM2; lanes 2 and 5 with thalamic tissue and lanes 3 and 6 with the cerebellum of two sCJDMM1-2 cases, respectively (T1 = PrPSc type 1; T2 = PrPSc type 2).
Figure 3
Examples of PK-resistant PrPSc distribution in various brain regions in sCJDMM1, sCJDMM2 and sCJDMM1-2 according to the criteria applied in this study. (A–C) Only the unglycosylated PrPSc isoform is shown in each box. (A) In sCJDMM1, PrPSc type 1 (20 kDa, arrow) but not PrPSc type 2 is detected in several brain regions after probing with mAb 1E4; PrPSc type 2 (T2) is shown at each edge of the gel as a control. (B) The PrPSc associated with sCJDMM2 (19 kDa, arrow) immunoreact with mAb 3F4. A PK-resistant PrPSc fragment of 18 kDa is often associated with PrPSc type 2 (18 kDa, arrow) (Pan et al., 2005); PrPSc type 1 (T1) is shown at each edge of the gel as control. (C) A case of sCJDMM1-2 with PrPSc types 1 and 2 co-occurring (20 and 19 kDa, arrows) in four brain regions (FC, VC, EC and CE) after incubation with 3F4. However, with 1E4 a 19 kDa band matching PrPSc type 2 was detected in the striatum (denoted by asterisk) and thalamus (denoted by double asterisk) even after loading one-half of the original amount used for PrPSc incubation with 3F4. FCS = frontal cortex (cx), superior gyrus; FCM = frontal cx, middle gyrus; FCMP = frontal cx, more posterior middle gyrus; VC = visual cx; OCI = occipital cx, inferior gyrus; EC = entorhinal cx; ST = striatum; TH = thalamus; PaqG = periaqueductal gray; Tct = tectum; PvG = periventricular grey; Olv = inferior olive; CE = cerebellum; T2 = PrPSc type 2; T1 = PrPSc type 1.
Figure 4
CSIs of PrPSc from each sCJDMM group of cases. (A) Representative WB of PK-resistant PrPSc from sCJDMM1, sCJDMM2 and sCJDMM1-2 probed with 3F4 at increasing molar (M) concentrations of GdnHCl and used for the CSI shown in (B). The case of sCJDMM1-2 illustrated here is case 6 of Table 3, and is identified with an asterisk in (B). The two arrows indicate the unglycosylated bands belonging to PrPSc type 1 (T1) and type 2 (T2) analysed for the sCJDMM1-2 of C. (B) GdnHCl molar amounts needed to render PK-sensitive one-half of the PrPSc, [GdnHCl]1/2, as determined with CSI performed with the whole PrPSc from sCJDMM1, sCJDMM2 and sCJDMM1-2 subtypes. PrPSc from sCJDMM1 (blue circles), sCJDMM1-2 (black diamonds) and sCJDMM2 (red triangles) show distinct [GdnHCl]1/2 values. The sCJDMM1-2 group appears to include two populations with a mean [GdnHCl]1/2 of 2.4 ± 0.2 and 1.7 ± 0.1, respectively. The [GdnHCl]1/2 correlated with the disease duration, which was 4.4 ± 2.8 months in the cases of the upper cluster and 7.8 ± 3.2 months in the lower. The lower cluster includes cases 4, 12, 13, 14 and 15 of Table 3. In 1 of the 7 sCJDMM1 cases and in patients 3 and 1 of the 13 cases of sCJDMM1-2, [GdnHCl]1/2 represents the mean of 2 or 3 brain regions. The disease duration of the seven sCJDMM1 cases spans from 1 month [(GdnHCl)1/2 = 2.54 M] to 24 months [(GdnHCl)1/2 = 2.51 M]. (C) [GdnHCl]1/2 of the unglycosylated PrPSc isoform from sCJDMM1 (blue circles), sCJDMM1-2 (black diamonds), sCJDMM2 (red triangles) and PrPSc types 1 and 2 from sCJDMM1 and sCJDMM2 mixed in vitro (black squares). The 17 tests from the sCJDMM1-2 cases are grouped as follows: tests of PrPSc types 1 or 2 present separately (alone), and of PrPSc types 1 and 2 co-existing in the same preparation but quantified separately by densitometric analysis of the corresponding unglycosylated band [see arrows in (A)] (co-exist). (D) CSI curves of the unglycosylated PrPSc isoform from the sCJDMM1-2 regions in which PrPSc type 2, co-existing with PrPSc type 1 in the same brain region, was recognized either by both mAb 3F4 and 1E4 (green line) (n = 7 regions) or exclusively by 1E4 (orange line) (n = 4 regions). The values of [GdnHCl]1/2 for each curve are indicated on the _X_-axis; (M) = molarity.
Figure 5
Spongiform profile of the sCJDMM patients. (A) Percent distribution of large vacuole spongiform degeneration in sCJDMM1, sCJDMM2 and sCJDMM1-2. Individual data points are the mean percentage of the surfaces occupied by large vacuole spongiform degeneration characteristic of sCJDMM2. Standard deviations are omitted for clarity. In the cerebellum, the bars refer to the percentage of the surface of the molecular layer occupied by the ‘fine’ type of spongiform degeneration characteristic of sCJDMM1. On average of all regions, sCJDMM1-2 had 3.2 times and statistically significant less large vacuole spongiform degeneration than sCJDMM2 (P < 0.05). No large vacuole spongiform degeneration was seen in sCJDMM1. The profiles of sCJDMM1-2 of long (≥8 months) and short (<8 months) durations were not significantly different (P > 0.05). The fine spongiform degeneration in the cerebellum of sCJDMM2 cases was significantly less (P < 0.05) than in the other subtypes. Statistical analyses were performed by ANOVA followed by the Dunn's multiple comparison post-test. (B) sCJDMM1 fine spongiform degeneration. (C) sCJDMM2 large vacuole spongiform degeneration. (D) Fine, punctuate (circle) and plaque-like (squares) patterns of PrP immunoreactivity in the cerebellum of the sCJDMM1-2. FC = frontal cortex; TC = temporal cortex; PC = parietal cortex; OC = occipital (visual) cortex; HI = hippocampus; EC = entorhinal cortex; BG = basal ganglia (putamen); TH = thalamus (anterior and mediodorsal nuclei); SN = substantia nigra.
Comment in
- Sporadic Creutzfeldt-Jakob disease: discrete subtypes or a spectrum of disease?
Head MW, Ironside JW. Head MW, et al. Brain. 2009 Oct;132(Pt 10):2627-9. doi: 10.1093/brain/awp225. Epub 2009 Sep 16. Brain. 2009. PMID: 19759201 No abstract available.
Similar articles
- Classification of sporadic Creutzfeldt-Jakob disease revisited.
Cali I, Castellani R, Yuan J, Al-Shekhlee A, Cohen ML, Xiao X, Moleres FJ, Parchi P, Zou WQ, Gambetti P. Cali I, et al. Brain. 2006 Sep;129(Pt 9):2266-77. doi: 10.1093/brain/awl224. Brain. 2006. PMID: 16923954 - Prions from Sporadic Creutzfeldt-Jakob Disease Patients Propagate as Strain Mixtures.
Cassard H, Huor A, Espinosa JC, Douet JY, Lugan S, Aron N, Vilette D, Delisle MB, Marín-Moreno A, Peran P, Beringue V, Torres JM, Ironside JW, Andreoletti O. Cassard H, et al. mBio. 2020 Jun 16;11(3):e00393-20. doi: 10.1128/mBio.00393-20. mBio. 2020. PMID: 32546613 Free PMC article. - Protease-sensitive conformers in broad spectrum of distinct PrPSc structures in sporadic Creutzfeldt-Jakob disease are indicator of progression rate.
Kim C, Haldiman T, Cohen Y, Chen W, Blevins J, Sy MS, Cohen M, Safar JG. Kim C, et al. PLoS Pathog. 2011 Sep;7(9):e1002242. doi: 10.1371/journal.ppat.1002242. Epub 2011 Sep 8. PLoS Pathog. 2011. PMID: 21931554 Free PMC article. - Creutzfeldt-Jakob disease.
Sikorska B, Knight R, Ironside JW, Liberski PP. Sikorska B, et al. Adv Exp Med Biol. 2012;724:76-90. doi: 10.1007/978-1-4614-0653-2_6. Adv Exp Med Biol. 2012. PMID: 22411235 Review. - Sporadic and familial CJD: classification and characterisation.
Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Gambetti P, et al. Br Med Bull. 2003;66:213-39. doi: 10.1093/bmb/66.1.213. Br Med Bull. 2003. PMID: 14522861 Review.
Cited by
- The Mutability of Yeast Prions.
King CY. King CY. Viruses. 2022 Oct 25;14(11):2337. doi: 10.3390/v14112337. Viruses. 2022. PMID: 36366434 Free PMC article. - Distinct pathological phenotypes of Creutzfeldt-Jakob disease in recipients of prion-contaminated growth hormone.
Cali I, Miller CJ, Parisi JE, Geschwind MD, Gambetti P, Schonberger LB. Cali I, et al. Acta Neuropathol Commun. 2015 Jun 25;3:37. doi: 10.1186/s40478-015-0214-2. Acta Neuropathol Commun. 2015. PMID: 26108478 Free PMC article. - A novel mechanism of phenotypic heterogeneity in Creutzfeldt-Jakob disease.
Nemani SK, Xiao X, Cali I, Cracco L, Puoti G, Nigro M, Lavrich J, Bharara Singh A, Appleby BS, Sim VL, Notari S, Surewicz WK, Gambetti P. Nemani SK, et al. Acta Neuropathol Commun. 2020 Jun 19;8(1):85. doi: 10.1186/s40478-020-00966-x. Acta Neuropathol Commun. 2020. PMID: 32560672 Free PMC article. - Clinical Use of Improved Diagnostic Testing for Detection of Prion Disease.
Figgie MP Jr, Appleby BS. Figgie MP Jr, et al. Viruses. 2021 Apr 28;13(5):789. doi: 10.3390/v13050789. Viruses. 2021. PMID: 33925126 Free PMC article. Review. - MM2-thalamic Creutzfeldt-Jakob disease: neuropathological, biochemical and transmission studies identify a distinctive prion strain.
Moda F, Suardi S, Di Fede G, Indaco A, Limido L, Vimercati C, Ruggerone M, Campagnani I, Langeveld J, Terruzzi A, Brambilla A, Zerbi P, Fociani P, Bishop MT, Will RG, Manson JC, Giaccone G, Tagliavini F. Moda F, et al. Brain Pathol. 2012 Sep;22(5):662-9. doi: 10.1111/j.1750-3639.2012.00572.x. Epub 2012 Feb 21. Brain Pathol. 2012. PMID: 22288561 Free PMC article.
References
- Cali I, Castellani R, Yuan J, Al-Shekhlee A, Cohen ML, Xiao X, et al. Classification of sporadic Creutzfeldt-Jakob disease revisited. Brain. 2006;129(Pt 9):2266–77. - PubMed
- Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and prion diseases. J Biol Chem. 1995;270:19173–80. - PubMed
- Collins SJ, Sanchez-Juan P, Masters CL, Klug GM, van Duijn C, Poleggi A, et al. Determinants of diagnostic investigation sensitivities across the clinical spectrum of sporadic Creutzfeldt-Jakob disease. Brain. 2006;129(Pt 9):2278–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials