CpG-depleted promoters harbor tissue-specific transcription factor binding signals--implications for motif overrepresentation analyses - PubMed (original) (raw)
CpG-depleted promoters harbor tissue-specific transcription factor binding signals--implications for motif overrepresentation analyses
Helge G Roider et al. Nucleic Acids Res. 2009 Oct.
Abstract
Motif overrepresentation analysis of proximal promoters is a common approach to characterize the regulatory properties of co-expressed sets of genes. Here we show that these approaches perform well on mammalian CpG-depleted promoter sets that regulate expression in terminally differentiated tissues such as liver and heart. In contrast, CpG-rich promoters show very little overrepresentation signal, even when associated with genes that display highly constrained spatiotemporal expression. For instance, while approximately 50% of heart specific genes possess CpG-rich promoters we find that the frequently observed enrichment of MEF2-binding sites upstream of heart-specific genes is solely due to contributions from CpG-depleted promoters. Similar results are obtained for all sets of tissue-specific genes indicating that CpG-rich and CpG-depleted promoters differ fundamentally in their distribution of regulatory inputs around the transcription start site. In order not to dilute the respective transcription factor binding signals, the two promoter types should thus be treated as separate sets in any motif overrepresentation analysis.
Figures
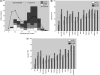
Figure 1.
(a) Bimodal CpG distribution across the promoters of the 50, 200 or 500 most heart-specific genes. The fraction of HCPs in the 200 and 500 gene sets is larger than expected based on the CpG content across all mouse promoters (the black line indicates the expected CpG distribution for random gene sets of size 500). (b) and (c) show the contribution of HCP genes to other tissue sets of indicated size based on EST or microarray data, respectively. The contribution of HCP promoters usually increases with gene set size and is >20% even for most sets of size 50. Sets with more than 200 genes often contain an excess of HCP genes compared to the fraction of 46% of HCPs across all Ensembl promoters (indicated by horizontal lines).
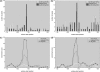
Figure 2.
Enrichment of high affinity sites for HNF1 and MEF2 near the TSS of liver- and muscle-specific genes with CpG-depleted promoters. (a) and (b) Sequence windows with highest affinity are preferentially located directly upstream of the TSS (red bars). This signal is due to sites in the CpG-depleted promoters as no preferential binding pattern is observed when restricting the analysis to CpG-rich promoters (compare blue and yellow bars for results from CpG-rich and CpG-depleted genes, respectively). (c) and (d) show the corresponding PASTAA enrichment scores (see ‘Materials and Methods’ section) for each sequence window as well as the separate sets of high and low CpG promoters.
Figure 3.
TF-binding affinity enrichment near the TSS of tissue-specific genes with either CpG-rich (a) or CpG-depleted promoters (b). The height of each bar corresponds to the PASTAA enrichment score of the most significant association that is found for the corresponding tissue. With the exception of testis, no significant enrichment signals are detected when analyzing the tissue sets containing the 500 most specific CpG-rich promoters. In contrast, enrichment peaks strongly for tissue-specific sets of 500 _P_-value matched LCP genes when computing TF-binding affinities for 200-bp windows directly upstream of the TSSs.
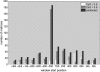
Figure 4.
TF targets have low average CpG content. Yellow and blue bars indicate the number of matrices whose target genes have an average CpG content ≥0.5 and <0.5, respectively. Red bars indicate the overall propensity to find the most significant association between matrices and any of the tissues at a particular window position. About a third of all matrices show the strongest association with any of the tissues when computing the binding affinities for the window ranging from −200 to 0 bp upstream of the TSS, indicating a strong location preference for the proximal promoter (see red bars). The target genes of the vast majority of matrices thereby have an average CpG content <0.5 (compare yellow and blue bars).
Similar articles
- Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters.
Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV. Oei SL, et al. Genomics. 2004 May;83(5):873-82. doi: 10.1016/j.ygeno.2003.11.001. Genomics. 2004. PMID: 15081116 - All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues.
Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C. Rozenberg JM, et al. BMC Genomics. 2008 Feb 5;9:67. doi: 10.1186/1471-2164-9-67. BMC Genomics. 2008. PMID: 18252004 Free PMC article. - Analysis of Pig Vomeronasal Receptor Type 1 (V1R) Promoter Region Reveals a Common Promoter Motif but Poor CpG Islands.
Dinka H, Le MT. Dinka H, et al. Anim Biotechnol. 2018;29(4):293-300. doi: 10.1080/10495398.2017.1383915. Epub 2017 Nov 9. Anim Biotechnol. 2018. PMID: 29120694 - Functional relevance of CpG island length for regulation of gene expression.
Elango N, Yi SV. Elango N, et al. Genetics. 2011 Apr;187(4):1077-83. doi: 10.1534/genetics.110.126094. Epub 2011 Feb 1. Genetics. 2011. PMID: 21288871 Free PMC article. - Screening of transcription factors with transcriptional initiation activity.
Zhang L, Yu H, Wang P, Ding Q, Wang Z. Zhang L, et al. Gene. 2013 Nov 15;531(1):64-70. doi: 10.1016/j.gene.2013.07.054. Epub 2013 Aug 9. Gene. 2013. PMID: 23933270
Cited by
- CpG island-mediated global gene regulatory modes in mouse embryonic stem cells.
Beck S, Lee BK, Rhee C, Song J, Woo AJ, Kim J. Beck S, et al. Nat Commun. 2014 Nov 18;5:5490. doi: 10.1038/ncomms6490. Nat Commun. 2014. PMID: 25405324 Free PMC article. - Developmental alterations in Huntington's disease neural cells and pharmacological rescue in cells and mice.
HD iPSC Consortium. HD iPSC Consortium. Nat Neurosci. 2017 May;20(5):648-660. doi: 10.1038/nn.4532. Epub 2017 Mar 20. Nat Neurosci. 2017. PMID: 28319609 Free PMC article. - DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients.
Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, Del Guerra S, Masini M, Bugliani M, Marchetti P, Cnop M, Eizirik DL, Fuks F. Volkmar M, et al. EMBO J. 2012 Mar 21;31(6):1405-26. doi: 10.1038/emboj.2011.503. Epub 2012 Jan 31. EMBO J. 2012. PMID: 22293752 Free PMC article. - A novel unbiased measure for motif co-occurrence predicts combinatorial regulation of transcription.
Vandenbon A, Kumagai Y, Akira S, Standley DM. Vandenbon A, et al. BMC Genomics. 2012;13 Suppl 7(Suppl 7):S11. doi: 10.1186/1471-2164-13-S7-S11. Epub 2012 Dec 13. BMC Genomics. 2012. PMID: 23282148 Free PMC article. - Chromatin and epigenetic features of long-range gene regulation.
Harmston N, Lenhard B. Harmston N, et al. Nucleic Acids Res. 2013 Aug;41(15):7185-99. doi: 10.1093/nar/gkt499. Epub 2013 Jun 13. Nucleic Acids Res. 2013. PMID: 23766291 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources