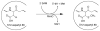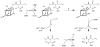Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily - PubMed (original) (raw)
Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily
Kyung-Hoon Lee et al. Biochemistry. 2009.
Abstract
RimO, encoded by the yliG gene in Escherichia coli, has been recently identified in vivo as the enzyme responsible for the attachment of a methylthio group on the beta-carbon of Asp88 of the small ribosomal protein S12 [Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1826-1831]. To date, it is the only enzyme known to catalyze methylthiolation of a protein substrate; the four other naturally occurring methylthio modifications have been observed on tRNA. All members of the methylthiotransferase (MTTase) family, to which RimO belongs, have been shown to contain the canonical CxxxCxxC motif in their primary structures that is typical of the radical S-adenosylmethionine (SAM) family of proteins. MiaB, the only characterized MTTase, and the enzyme experimentally shown to be responsible for methylthiolation of N(6)-isopentenyladenosine of tRNA in E. coli and Thermotoga maritima, has been demonstrated to harbor two distinct [4Fe-4S] clusters. Herein, we report in vitro biochemical and spectroscopic characterization of RimO. We show by analytical and spectroscopic methods that RimO, overproduced in E. coli in the presence of iron-sulfur cluster biosynthesis proteins from Azotobacter vinelandii, contains one [4Fe-4S](2+) cluster. Reconstitution of this form of RimO (RimO(rcn)) with (57)Fe and sodium sulfide results in a protein that contains two [4Fe-4S](2+) clusters, similar to MiaB. We also show by mass spectrometry that RimO(rcn) catalyzes the attachment of a methylthio group to a peptide substrate analogue that mimics the loop structure bearing aspartyl 88 of the S12 ribosomal protein from E. coli. Kinetic analysis of this reaction shows that the activity of RimO(rcn) in the presence of the substrate analogue does not support a complete turnover. We discuss the possible requirement for an assembled ribosome for fully active RimO in vitro. Our findings are consistent with those of other enzymes that catalyze sulfur insertion, such as biotin synthase, lipoyl synthase, and MiaB.
Figures
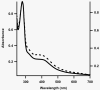
Figure 1
UV-visible spectra of RimOai (solid line, left Y-axis) and RimOrcn (dashed line, right Y-axis). Sample concentrations were 9.5 μM for RimOai and 5.5 μM for RimOrcn. A280/A410 ratios are 4.2 and 2.9 for RimOai and RimOrcn, respectively.
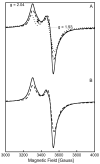
Figure 2
X-band EPR spectra of (A) RimOai and (B) RimOai reduced with sodium dithionite. The spectra in (A) and (B, solid line) were recorded at a temperature of 14 K, while the spectrum in (B, dashed line) was recorded at a temperature of 64 K. The arrow in spectrum (B, solid line) is pointing to a g = 2.0 signal resulting from a cavity contaminant. In both panels, the concentration of the protein sample was 0.45 mM. In (B), the sample was reduced by the addition of 2 mM sodium dithionite at room temperature for ∼2 min before freezing it in liquid nitrogen. Conditions of measurements were as follows: microwave power, 1 mW; receiver gain, 2 × 104; modulation amplitude, 10 G; microwave frequency, 9.51 GHz.
Figure 3
4.2-K Mössbauer spectra of RimO. All spectra were collected in an external 53-mT magnetic field except for (B), which was collected in a 6-T magnetic field oriented parallel to the γ-beam. (A) RimOai (hashed marks) and simulation with two quadrupole doublets (solid line) with the following parameters: _δ_1 = 0.43 mm/s and _ΔEQ,_1 = 1.07 mm/s (90% intensity) and _δ_2 = 0.29 mm/s and _ΔEQ,_2 = 0.49 mm/s (10% intensity). The arrow is pointing to a line at ∼0.5 mm/s. (B) RimOai (hashed marks) and spin Hamiltonian simulations (solid line) using the values for δ and ΔEQ from (A) and the asymmetry parameters η1 = 0 and η2 = 1. (C) RimOai in the presence of SAM (hashed marks) and in the presence of SAM and P1 (solid line). (D) Difference spectra between the hashed marked spectra (A)-(C) (hashed marks) and simulation of the difference spectrum with three quadrupole doublets (solid line) with the following parameters: _δ_1 = 0.43 mm/s and _ΔEQ,_1 = 1.07 mm/s (32% downwards), _δ_2 = 0.70 mm/s and _ΔEQ,_2 = 1.24 mm/s (16% upwards), and _δ_3 = 0.37 mm/s and _ΔEQ,_3 = 0.81 mm/s (16% upwards). (E) RimOrcn (hashed marks) and simulation of the [4Fe-4S]2+ component with one quadrupole doublet (solid line) with the following parameters: _δ_1 = 0.43 mm/s and _ΔEQ,_1 = 1.12 mm/s (62% intensity). (F) RimOai in the presence of SAM (hashed marks) and in the presence of SAM and P1 (solid line). (G) Comparison of the difference spectra obtained for binding of SAM in RimOai and RimOrcn. The hashed marks correspond to the difference spectrum (E) – (F), and the solid line to (A) – (C). The concentrations of RimOai and RimOrcn were 0.45 mM and 0.26 mM, respectively. SAM was used at a concentration of 3.5 mM. P1 was used at a concentration of 3.7 mM.
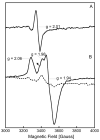
Figure 4
X-band EPR spectra of (A) reduced RimOrcn (solid line), reduced RimOai (dotted line), and reduced RimOrcn in the presence of SAM (dashed line); (B) reduced RimOrcn (solid line) and reduced RimOrcn in the presence of SAM and P1 (dashed line). The concentrations of RimOai and RimOrcn were 0.45 mM and 0.26 mM, respectively. SAM was used at a concentration of 3.5 mM, while P1 was used at a concentration of 3.7 mM. Each RimO sample was reduced by the addition of 2 mM sodium dithionite at room temperature for ∼2 min before freezing in liquid nitrogen. The solid-line spectra in (A) and (B) display g-values ≈ 2.04 (_g_‖) and ≈ 1.93 (_g_⊥). The dashed-line spectra in (A) and (B) display g-values ≈ 2.04 (_g_‖) and ≈ 1.93 (_g_⊥). The dotted-line spectrum in (A) displays g-values ≈ 2.05 (_g_‖) and ≈ 1.93 (_g_⊥). In this same spectrum, the arrow is pointing to a g ≈ 2.0 signal resulting from a cavity contaminant. Different scaling has been used for different spectra: × 1.0 for solid-line spectra in (A) and (B); × 5.0 for dotted-line spectrum in (A); × 2.4 for dashed-line spectrum in (A); and × 2.9 for dashed-line spectrum in (B). Conditions of measurements were as follows: microwave power, 20 mW; temperature, 14 K; receiver gain, 2 × 104; modulation amplitude, 10 G; microwave frequency, 9.51 GHz.
Figure 5
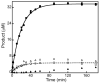
Time courses for the formation of 5′-dA and SAH in the reaction of RimOrcn with peptide-substrate analogs P1 or P2 at 37 °C. Closed circles (●) describe formation of 5′-dA and open circles (○) describe formation of SAH in the presence of P1. The solid and dashed lines are fits to a single first-order kinetic equation. The kinetic parameters obtained from these two fits are A = 32.9 ± 0.4 μM, k = 0.050 ± 0.001 min-1, and m = 0.91 ± 0.29 μM for formation of 5′-dA (solid line) and A = 4.1 ± 0.1 μM, k = 0.058 ± 0.006 min-1, and m = 0.63 ± 0.12 μM for formation of SAH (dashed line), where A is the amplitude, k is the rate constant at 37 °C, and m is the intercept. Closed squares (■) describe the formation of 5′-dA and open squares (□) describe the formation of SAH in the presence of P1 and RNA oligo. Closed triangles (▲) describe the formation of 5′-dA and open triangles (△) describe the formation of SAH in the presence of P2. The reaction was carried out as described in Materials and Methods and contained 100 μM of RimOrcn, 700 μM SAM, 300 μM peptide, 50 mM Na-HEPES (pH 7.5), 2 mM dithionite, 1 mM tryptophan, and 175 μM RNA oligo where indicated.
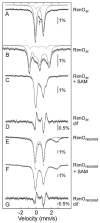
Figure 6
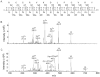
ESI-MS/MS spectra of the 4+ ion of P1. (A) b and y fragment ions and their expected masses for P1; (B) unmodified P1 (m/z 369.17) at t = 0 min; (C) modified P1 (m/z 380.9) at t = 15 min contains both the b72+ and b82+ + 47 ions, indicative of methylthiolation on the Asp residue. The reaction mixture contained 100 μM RimOrcn, 700 μM SAM, 300 μM P1, 50 mM Na-HEPES (pH 7.5), and 2 mM dithionite. Sample preparation is as discussed in Materials and Methods.
Scheme 1
Reaction catalyzed by E. coli RimO, the methylthiolation of aspartyl 88 of protein S12 of the ribosome
Scheme 2
Working mechanistic hypothesis for the reaction catalyzed by E. coli RimO. The [4Fe–4S] cluster shown is that which is predicted to be coordinated by C17, C53, and C82. This cluster has an overall charge of +2, indicating the presence of formally 2 Fe3+ and 2 Fe2+ ions in its resting state. Two scenarios are shown for methylthiolation. In pathway 1, sulfur functionalization by a bridging μ-sulfido ligand of the cluster leads to cluster degradation. The resulting thiol-containing intermediate is methylated in a subsequent step that might potentially take place at a different location. In pathway 2, methylation of the attached sulfur atom takes place while it is still part of the Fe/S cluster, which subsequently leads to cluster degradation.
Similar articles
- Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase.
Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M. Arragain S, et al. J Biol Chem. 2010 Feb 19;285(8):5792-801. doi: 10.1074/jbc.M109.065516. Epub 2009 Dec 9. J Biol Chem. 2010. PMID: 20007320 Free PMC article. - Stereochemical Course of the Reaction Catalyzed by RimO, a Radical SAM Methylthiotransferase.
Landgraf BJ, Booker SJ. Landgraf BJ, et al. J Am Chem Soc. 2016 Mar 9;138(9):2889-92. doi: 10.1021/jacs.5b11035. Epub 2016 Feb 25. J Am Chem Soc. 2016. PMID: 26871608 - Redox Behavior of the S-Adenosylmethionine (SAM)-Binding Fe-S Cluster in Methylthiotransferase RimO, toward Understanding Dual SAM Activity.
Molle T, Moreau Y, Clemancey M, Forouhar F, Ravanat JL, Duraffourg N, Fourmond V, Latour JM, Gambarelli S, Mulliez E, Atta M. Molle T, et al. Biochemistry. 2016 Oct 18;55(41):5798-5808. doi: 10.1021/acs.biochem.6b00597. Epub 2016 Oct 6. Biochemistry. 2016. PMID: 27677419 - The methylthiolation reaction mediated by the Radical-SAM enzymes.
Atta M, Arragain S, Fontecave M, Mulliez E, Hunt JF, Luff JD, Forouhar F. Atta M, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1223-30. doi: 10.1016/j.bbapap.2011.11.007. Epub 2011 Dec 7. Biochim Biophys Acta. 2012. PMID: 22178611 Free PMC article. Review. - The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase.
Jarrett JT. Jarrett JT. Arch Biochem Biophys. 2005 Jan 1;433(1):312-21. doi: 10.1016/j.abb.2004.10.003. Arch Biochem Biophys. 2005. PMID: 15581586 Review.
Cited by
- Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
Lachowicz JC, Lennox-Hvenekilde D, Myling-Petersen N, Salomonsen B, Verkleij G, Acevedo-Rocha CG, Caddell B, Gronenberg LS, Almo SC, Sommer MOA, Genee HJ, Grove TL. Lachowicz JC, et al. J Am Chem Soc. 2024 Jan 24;146(3):1860-1873. doi: 10.1021/jacs.3c05481. Epub 2024 Jan 12. J Am Chem Soc. 2024. PMID: 38215281 - Essentiality of the Escherichia coli YgfZ Protein for the In Vivo Thiomethylation of Ribosomal Protein S12 by the RimO Enzyme.
Lund T, Kulkova MY, Jersie-Christensen R, Atlung T. Lund T, et al. Int J Mol Sci. 2023 Mar 1;24(5):4728. doi: 10.3390/ijms24054728. Int J Mol Sci. 2023. PMID: 36902159 Free PMC article. - ThnL, a B12-dependent radical _S_-adenosylmethionine enzyme, catalyzes thioether bond formation in carbapenem biosynthesis.
Sinner EK, Li R, Marous DR, Townsend CA. Sinner EK, et al. Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2206494119. doi: 10.1073/pnas.2206494119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969793 Free PMC article. - Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Shi YM, Hirschmann M, Shi YN, Ahmed S, Abebew D, Tobias NJ, Grün P, Crames JJ, Pöschel L, Kuttenlochner W, Richter C, Herrmann J, Müller R, Thanwisai A, Pidot SJ, Stinear TP, Groll M, Kim Y, Bode HB. Shi YM, et al. Nat Chem. 2022 Jun;14(6):701-712. doi: 10.1038/s41557-022-00923-2. Epub 2022 Apr 25. Nat Chem. 2022. PMID: 35469007 Free PMC article. - Biochemical Approaches to Probe the Role of the Auxiliary Iron-Sulfur Cluster of Lipoyl Synthase from Mycobacterium Tuberculosis.
Jeyachandran VR, Pendyala JV, McCarthy EL, Boal AK, Booker SJ. Jeyachandran VR, et al. Methods Mol Biol. 2021;2353:307-332. doi: 10.1007/978-1-0716-1605-5_16. Methods Mol Biol. 2021. PMID: 34292556
References
- Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. - PubMed
- Pierrel F, Björk GR, Fontecave M, Atta M. Enzymatic modification of tRNAs: MiaB is an iron–sulfur protein. J Biol Chem. 2002;277:13367–13370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous